Abstract
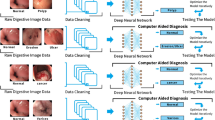
In recent years, artificial intelligence and its tools are demonstrated enough potential for analyzing medical images. Several deep learning models have been proposed in previous studies for gastrointestinal (GI) tract like ulcers, polyps, bleeding, and other lesions. Hand-operated investigation of these lesions requires time, cost, and an expert physician. Automatic detection and classification of GI tract lesions are vital because misdiagnosis of them can affect the quality of human life. In our study, an effective model is proposed for a GI tract classification with the best performance. The proposed method’s main aim is to classify GI tract lesions precisely from endoscopic video frames automatically. The different scenarios are designed, assessed, and compared by implementing 5-fold cross-validation on the KVASIR V1 dataset to achieve this aim. This dataset includes anatomical landmarks (pylorus, z-line, and cecum), pathological findings (esophagitis, ulcerative colitis, and polyp), and polyp removals (dyed lifted polyps, and dyed resection margins) as output classes. Each class includes 500 images, and an image’s resolution varies from 750 × 576 to 1920 × 1072 pixels. These first and second scenarios are based on deep neural networks (DNNs). However, in the first scenario, a novel approach is proposed for visualizing 2-D data maps from features extracted from the convolutional auto-encoder (CAE). The last one is schemed based on pre-trained convolutional neural networks (CNNs). The experimental results illustrate the average accuracy of the first, second, and third scenarios is 99.87 ± 0.001, 92.07 ± 0.086, and 90.55 ± 0.111, respectively. The first scenario outperforms the compared ones with an average accuracy of 99.87 ± 0.001 and an AUC of 100.00 ± 0.000.







Similar content being viewed by others
Data availability
We use KVASIR V1 dataset in this study for designing and examining our proposed which is publicly available at https://datasets.simula.no/kvasir/.
References
Ahmad J, Muhammad K, Lee MY, Baik SW (2017) Endoscopic image classification and retrieval using clustered convolutional features, (in Eng). J Med Syst 41(12):196. https://doi.org/10.1007/s10916-017-0836-y
Asperti A, Mastronardo C (2017) The effectiveness of data augmentation for detection of gastrointestinal diseases from endoscopical images. arXiv preprint arXiv:1712 03689. https://doi.org/10.1016/j.compmedimag.2020.101852
Borgli H et al (2020) HyperKvasir, a comprehensive multi-class image and video dataset for gastrointestinal endoscopy. Sci Data 7(1):1–14. https://doi.org/10.1038/s41597-020-00622-y
Caroppo A, Leone A, Siciliano P (2021) Deep transfer learning approaches for bleeding detection in endoscopy images. Comput Med Imaging Graph 88:101852. https://doi.org/10.1016/j.compmedimag.2020.101852
Chauhan NK, Singh K (2018) A review on conventional machine learning vs deep learning. In: 2018 International Conference on Computing, Power and Communication Technologies (GUCON), pp 347–352, https://doi.org/10.1109/GUCON.2018.8675097
Elhami G, Weber RM (2019) Audio feature extraction with convolutional neural autoencoders with application to voice conversion. Conference: infoscience
Ghosh T, Chakareski J (2021) Deep transfer learning for automated intestinal bleeding detection in Capsule Endoscopy Imaging. J Digit Imaging. https://doi.org/10.1007/s10278-021-00428-3
Guo X, Yuan Y (2020) Semi-supervised WCE image classification with adaptive aggregated attention. Med Image Anal 64:101733. https://doi.org/10.1016/j.media.2020.101733
Han J, Kamber M, Pei J (2011) Data mining concepts and techniques, 3rd edn. The Morgan Kaufmann Series in Data Management Systems 5(4):83–124. https://doi.org/10.1016/C2009-0-61819-5
Hasan MM, Hossain MM, Mia S, Ahammad MS, Rahman MM (2022) A combined approach of non-subsampled contourlet transform and convolutional neural network to detect gastrointestinal polyp. Multimedia Tools Appl 81(7):9949–9968. https://doi.org/10.1007/s11042-022-12250-2
Heidari M, Mirniaharikandehei S, Khuzani AZ, Danala G, Qiu Y, Zheng B (2020) Improving the performance of CNN to predict the likelihood of COVID-19 using chest X-ray images with preprocessing algorithms. Int J Med Inform 144:104284. https://doi.org/10.1016/j.ijmedinf.2020.104284
Hu H et al (2021) Content-based gastric image retrieval using convolutional neural networks. Int J Imaging Syst Technol 31(1):439–449. https://doi.org/10.1002/ima.22470
Hwang M et al (2020) An automated detection system for colonoscopy images using a dual encoder-decoder model, (in Eng). Comput Med Imaging Graph 84:101763. https://doi.org/10.1016/j.compmedimag.2020.101763
Jain S et al (2021) A deep CNN model for anomaly detection and localization in wireless capsule endoscopy images. Comput Biol Med 137:104789. https://doi.org/10.1016/j.compbiomed.2021.104789
Jha D et al (2020) Kvasir-seg: A segmented polyp dataset. In: International Conference on Multimedia Modeling, 2020. Springer, Berlin, pp 451–462. https://doi.org/10.1007/978-3-030-37734-2_37
Jia X, Meng MQ (2017) Gastrointestinal bleeding detection in wireless capsule endoscopy images using handcrafted and CNN features. In: 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 11–15 July 2017, pp 3154–3157, https://doi.org/10.1109/EMBC.2017.8037526
Khan MA et al (2022) GestroNet: a framework of saliency estimation and optimal deep learning features based gastrointestinal diseases detection and classification. Diagnostics 12(11):2718. [Online]. Available: https://www.mdpi.com/2075-4418/12/11/2718
Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv preprint arXiv: 1412.6980. https://doi.org/10.48550/arXiv.1412.6980
Leung WK, Cheung KS, Li B, Law SYK, Lui TKL (2021) Applications of machine learning models in the prediction of gastric cancer risk in patients after Helicobacter pylori eradication. Aliment Pharmacol Ther 53(8):864–872. https://doi.org/10.1111/apt.16272
Li L et al (2020) Multi-task deep learning for fine-grained classification and grading in breast cancer histopathological images. Multimedia Tools Appl 79:14509–14528. https://doi.org/10.1007/s11042-018-6970-9
Maggipinto M, Masiero C, Beghi A, Susto GA (2018) A convolutional autoencoder approach for feature extraction in virtual metrology. Procedia Manuf 17:126–133. https://doi.org/10.1016/j.promfg.2018.10.023
McClelland JL, Rumelhart DE, Group PR (1986) Parallel distributed processing. MIT Press, Cambridge. https://doi.org/10.7551/mitpress/5236.001.0001
Mohapatra S, Nayak J, Mishra M, Pati GK, Naik B, Swarnkar T (2021) Wavelet transform and deep convolutional neural network-based smart healthcare system for gastrointestinal disease detection, (in Eng). Interdiscip Sci. https://doi.org/10.1007/s12539-021-00417-8
Nair V, Hinton GE (2010) Rectified linear units improve restricted Boltzmann machines. In: ICML. https://dl.acm.org/doi/10.5555/3104322.3104425
Owais M, Arsalan M, Choi J, Mahmood T, Park KR (2019) Artificial intelligence-based classification of multiple gastrointestinal diseases using endoscopy videos for clinical diagnosis. J Clin Med 8(7):986. https://doi.org/10.3390/jcm8070986
Öztürk Ş, Özkaya U (2020) Gastrointestinal tract classification using improved LSTM based CNN. Multimedia Tools Appl 79(39):28825–28840. https://doi.org/10.1007/s11042-020-09468-3
Pannu HS, Ahuja S, Dang N, Soni S, Malhi AK (2020) Deep learning based image classification for intestinal hemorrhage. Multimedia Tools Appl 79:21941–21966. https://doi.org/10.1007/s11042-020-08905-7
Pogorelov K et al (2017) KVASIR: a Multi-Class Image dataset for computer aided gastrointestinal disease detection. https://doi.org/10.1145/3193289
Ponnusamy R, Sathiamoorthy S (2019) Prediction of esophagitis and Z-line from wireless capsule endoscopy images using fusion of low-level features. Int J Recent Technol Eng (IJRTE) 8(3):6024–6028. https://doi.org/10.35940/ijrte.C5568.098319
Raksasat R et al (2021) Accurate surface ultraviolet radiation forecasting for clinical applications with deep neural network. Sci Rep 11(1):5031. https://doi.org/10.1038/s41598-021-84396-2
Rau A et al (2019) Implicit domain adaptation with conditional generative adversarial networks for depth prediction in endoscopy. Int J Comput Assist Radiol Surg 14(7):1167–1176. https://doi.org/10.1007/s11548-019-01962-w
Safarov S, Whangbo TK (2021) A-denseunet: Adaptive densely connected unet for polyp segmentation in colonoscopy images with atrous convolution. Sensors 21(4):1–15, Art no. 1441. https://doi.org/10.3390/s21041441
Sokolova M, Lapalme G (2009) A systematic analysis of performance measures for classification tasks. Inf Process Manag 45(4):427–437. https://doi.org/10.1016/j.ipm.2009.03.002
Stone M (1974) Cross-validatory choice and assessment of statistical predictions. J R Stat Soc: Ser B (Methodological) 36(2):111–133. https://doi.org/10.1111/j.2517-6161.1974.tb00994.x
Sung H et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. https://doi.org/10.3322/caac.21660
Vieira PM, Freitas NR, Valente J, Vaz IF, Rolanda C, Lima CS (2020) Automatic detection of small bowel tumors in wireless capsule endoscopy images using ensemble learning. Med Phys 47(1):52–63. https://doi.org/10.1002/mp.13709
Xing X, Yuan Y, Meng MQH (2020) Zoom in lesions for better diagnosis: attention guided deformation network for WCE image classification. IEEE Trans Med Imaging 39(12):4047–4059. https://doi.org/10.1109/TMI.2020.3010102
Yuan Y, Meng MQH (2017) Deep learning for polyp recognition in wireless capsule endoscopy images. Med Phys 44(4):1379–1389. https://doi.org/10.1002/mp.12147
Zhang C, Zhang N, Wang D, Cao Y, Liu B (2020) Artifact detection in endoscopic video with deep convolutional neural networks. In: 2020 Second International Conference on Transdisciplinary AI (TransAI), pp 1–8. https://doi.org/10.1109/TransAI49837.2020.00007
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nezhad, S.A., Khatibi, T. & Sohrabi, M. Combining CNNs and 2-D visualization method for GI tract lesions classification. Multimed Tools Appl 83, 15825–15844 (2024). https://doi.org/10.1007/s11042-023-15347-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15347-4