Abstract
Diabetes is one of the most common and serious diseases affecting human health. Early diagnosis and treatment are vital to prevent or delay complications related to diabetes. An automated diabetes detection system assists physicians in the early diagnosis of the disease and reduces complications by providing fast and precise results. This study aims to introduce a technique based on a combination of multiple linear regression (MLR), random forest (RF), and XGBoost (XG) to diagnose diabetes from questionnaire data. MLR-RF algorithm is used for feature selection, and XG is used for classification in the proposed system. The dataset is the diabetic hospital data in Sylhet, Bangladesh. It contains 520 instances, including 320 diabetics and 200 control instances. The performance of the classifiers is measured concerning accuracy (ACC), precision (PPV), recall (SEN, sensitivity), F1 score (F1), and the area under the receiver-operating-characteristic curve (AUC). The results show that the proposed system achieves an accuracy of 99.2%, an AUC of 99.3%, and a prediction time of 0.04825 seconds. The feature selection method improves the prediction time, although it does not affect the accuracy of the four compared classifiers. The results of this study are quite reasonable and successful when compared with other studies. The proposed method can be used as an auxiliary tool in diagnosing diabetes.
Similar content being viewed by others
1 Introduction
Diabetes is a metabolic disorder characterized by high blood sugar levels that causes severe damage to the nerves, heart, eyes, kidneys, and blood vessels. Approximately 422 million people worldwide, mostly in low- and middle-income countries, have diabetes, and diabetes is the direct cause of more than 1.6 million deaths each year [53].
Diabetes is expected to increase significantly over the next decades [35]. Many studies have shown a higher susceptibility to certain infectious diseases in people with diabetes, possibly due to the dysregulated immune system [23]. It is reported that diabetes and plasma glucose levels in SARS patients are independent predictors of morbidity and mortality [58]. As with flu-related mortality [14], diabetes is a significant risk factor for the negative consequences of COVID-19. COVID-19 has recently become one of the serious and acute diseases that has spread all over the world, having a serious impact on the health system and the overall global economy [25, 45]. Mortality rates from pneumonia in people with diabetes aged 75 years and over in Hong Kong currently exceed death rates from cardiovascular disease and cancer in this age group [3]. A study conducted in Wuhan, China, revealed that 32% of 41 people infected with COVID-19 had underlying diseases and 20% had diabetes [52]. Guo et al. (2020) claimed that diabetes should be seen as a risk factor for the rapid progression and poor prognosis of COVID-19 rapid progression and poor prognosis [17]. Diabetic patients may be at high risk for COVID-19. Providing care to people with diabetes is an essential part of the effort [3].
The current diagnostic criteria used for the diagnosis of diabetes have been in place globally for almost a decade [54]. Plasma glucose measurement is one of the clinical diagnostic tests and continues to be the basis of diagnostic criteria. For this test, blood samples taken from the subjects are transported to the laboratories. Since blood cells can continue to metabolize glucose, the duration of blood glucose determination after blood is drawn is important [8]. Factors such as blood specimen collection, evaluation time, and instrumental errors can affect laboratory analysis results.
In the healthcare system, with the contribution of artificial intelligence-based systems, cancer cells [38], liver disorder [20], tumor detection [46], heart disease [2], breast cancer [16, 24, 26], COVID-19 [27, 28, 40, 44], and diabetes [1, 18, 21], great progress has been made for automatic detection of diseases. It is important to start early treatment with automatic early detection of diseases to slow down the disease process. Serious damages, morbidity, and mortality caused by diabetes can be reduced by early diagnosis and treatment. Therefore, my main focus is to develop an intelligent machine learning system that can detect and classify this disease. In recent years, researchers have provided intelligent diagnostic systems for diabetes detection to assist physicians. Kandhasamy and Balamurali (2015) compared four machine learning algorithms, including k-NN, support vector machines, random forest, and J48 decision tree, to predict diabetes mellitus [33]. Both k-NN (k = 1) and random forest, with 100% accuracy, were the best performing algorithms. A comparative study of Naive Bayes, ID3, random forest, and AdaBoost reported that random forest outperformed the other algorithms at classifying diabetic patients [56], with 85% accuracy. Paul and Choubey (2017) proposed a hybrid algorithm using a radial basis function network (RBFN) and a genetic algorithm (GA) [43]. The findings were that the hybrid method outperformed RBFN. Zou et al. (2018) used principal component analysis (PCA) and minimum redundancy maximum relevance (mRME) to reduce dimensionality [59]. They showed that the prediction could reach the highest accuracy with the random forest (ACC = 0.8084). Wu et al. (2018) have proposed a novel model based on data mining techniques (improved K-means algorithm and logistic regression) for predicting type 2 diabetes mellitus [55]. The model’s accuracy was up to 95.42%. Ayon ve Islam (2019), presented a deep learning network to automatically detect diabetes [1]. The dataset included a total of 768 samples, of which 268 were diabetic. The proposed system obtained 98.35% accuracy, an F1 score of 98, and an MCC of 97 for five-fold cross-validation. Islam et al. (2020) have analyzed the dataset (of this study) with the Naive Bayes Algorithm, Logistic Regression Algorithm, and Random [29]. They reported that the random forest using 16 features at its input reached the best result with an accuracy rate of 99%. Tigga and Garg (2020) conducted tests on the data set collected with 18 questions about diabetes to predict the risk of diabetes mellitus type 2 [49]. They applied different classification methods to the data and reported that the most successful accuracy rate was random forest, with 94.10%. Naz and Ahuja (2020) tested PIMA Indian diabetic dataset for diagnosis of diabetes disease using Naive Bayes (NB), Decision Tree (DT), Deep Learning (DL), and Artificial Neural Network (ANN) classifiers which give comparable performance [41]. The accuracy achieved by these classifiers is in the range of 90–98%. The DL provided the best results with an accuracy rate of 98.07%. Gupta et al. (2021) proposed an approach as logistic regression which is tested over a diabetic clinical dataset (DCA) which has shown better generalization performance compared to the other 14 classification techniques [18]. This dataset consists of 174 instances and 10 attributes, where a total of 61 nondiabetic and 113 diabetic patients. An accuracy of 94.59% was obtained for the diagnosis of diabetes. Gourisaria et al. (2022) used various deep learning, machine learning, and data dimensionality reduction techniques on two datasets to detect diabetes [15]. In their test on the second (University of California, Irvine) dataset, they achieved 99.2% accuracy with the Random Forest classifier using 16 features. When the dimensionality of the same dataset was reduced using linear discriminant analysis and principal component analysis, the accuracy dropped to 96.3 and 95.6, respectively.
In recent years, various methods have been proposed for diabetes prediction, including k-NN [33], LR [18], DL [1, 41], and RF [15, 29, 49, 56, 59]. However, these methods suffer from one or more of the following shortcomings: First, most of them do not test for statistical significance between the independent and dependent variables. This may create a trust problem regarding the use of variables used in the inputs of the classifiers. Second, some of them use more input data to increase the accuracy of the classifications. In this way, the response time of the classifier model increases. Third, most studies require various blood values to predict diabetes. The procedure for obtaining blood data is not easy and time-consuming. In this study, I propose a model that tests the reliability of independent variables created without blood data with a statistical significance test and requires a shorter time calculation with fewer features with a feature selection method. As far as I know, there is no study in the literature showing the use of MLR-RF and XGBoost as feature selection and classifier in diabetes prediction.
This paper proposes a novel method for predicting early-stage diabetes based on a data-driven. With the proposed method, the need for blood tests can be reduced by using direct questionnaires for diagnosing early-stage diabetes. The major contribution of this study is to find out a hybrid prediction model based on a machine learning approach with high prediction accuracy, low memory usage, and low computation time.
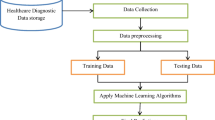
The proposed predictive model strategy based on MLR-RF-XG is shown in Fig. 1. The remainder of this paper is organized as follows: The materials and estimation methodology are described in Chapter 2. Experimental results, comparisons, and discussions are presented in Chapter 3. Finally, the conclusions are summarized in Chapter 4.
2 Materials and methodology
2.1 Datasets
The dataset used in this study was collected from patients of Sylhet Diabetes Hospital in Bangladesh using questionnaires. It was obtained from the publicly available web link https://archive.ics.uci.edu/ml/datasets/Early+stage+diabetes+risk+prediction+dataset [51]. The dataset format is CSV and constitutes 520 samples in which there are 200 “non-diabetes” and 320 “diabetes”. Each sample consists of gender, age, polydipsia, polyuria, weakness, visual blurring, sudden weight loss, genital thrush, itching, polyphagia, obesity, irritability, partial paresis, delayed healing, alopecia, and muscle stiffness.
2.2 Methodology
The proposed approach for detecting the risk of diabetes consists of five main stages: (1) descriptive statistics, (2) testing the validity of variables, (3) feature selection and reduction, (4) classification using all variables or using variables selected by RF, and (5) performance assessment and comparison. These stages are separated by a dotted line in Fig. 1.
In the first part of the study, the general descriptive statistics about independent variables were detailed. Then, MLR was used to test the validity of the variables. Third, the most significant risk factors that could cause diabetes with RF were identified. This step removes features that contain redundant information. A 16-dimensional feature vector is reduced to 9 with this strategy. Fourth, eight different tests are conducted using two different input sets and four different classifier algorithms for the classification process. Finally, the classifiers’ performances are evaluated and compared.
This study adopts a comparative approach between two different input set analysis frameworks, i.e., models I and II. Model I uses all independent variables as classifier inputs, while Model II adopts variables selected by MLR-RF. The rest of the framework is the same for both methods, and four machine learning techniques are named random forest (RF), k-nearest neighbor (k-NN), gradient boosting (GB), and XGBoost (XG) are compared.
2.2.1 Test of validity
Multiple linear regression (MLR)
MLR is a widely used regression analysis type [12]. The goal of multiple linear regression (MLR) is to describe data and model the linear relationship between the independent variables and one dependent variable. The relationship between dependent and independent variables focuses on understanding the effect of inputs on the output parameter.
The MLR method can be used to eliminate insignificant variables from the models using p value statistics. The p value is used to measure statistical significance in the context of null hypothesis testing. The smaller the p value, the more likely the findings will be valid. The p value for the 95% confidence level is 0.05. If the p value is less than 0.05, it indicates that the observed effect is not due to random changes [11]. A p value less than 0.05 for the 95% confidence level was considered statistically significant in this study.
2.2.2 Classifiers
This research aims to develop an optimal classification model that can be used for accurately detecting diabetes. It is also to determine the best model. The performances of four different models (k-Nearest Neighbors, gradient boosting, random forest, and XGBoost) were compared to achieve the best model.
K-nearest neighbors (k-NN)
The k-NN algorithm is an essential and straightforward data mining technique. It is especially useful in pattern recognition [10]. The main idea is that if a point to be estimated is in the same category as its nearest neighbors in the training set, it can be concluded that this point also has the same characteristics and attributes. The nearest neighbors in the training set affect the performance of the prediction results. Estimating a new point is about the values of the k-NN [7]. The k-NN algorithm has been widely used in many fields such as outlier detection, regression, and pattern recognition [42].
Random forest (RF)
Random forest (RF) can be defined as a collection of tree-type classifiers. Most datasets can contain multidimensional features with many irrelevant features. For classifier models, these irrelevant variables degrade classifier performance. Feature selection increases the success rate of the classifier. The random forest algorithm uses simple probability to select the strongest features for its inputs. Breiman (2001) formulated the RF algorithm using subsets of sample data and mapping a random sample of feature subspaces by constructing multiple decision trees [4, 30].
RF is an algorithm that scales well to large datasets and is resistant to irrelevant features. It also improves performance by reducing variance [37].
Gradient boosting (GB)
Gradient Boosting is a powerful technique that gives successful results in many practical applications [6, 22, 32, 48, 50]. GB discards all the weak predictors and chooses the stronger ones. It is an improved version of the decision tree where each successor is analyzed comparatively to create the best satisfactory structure of the tree using gain calculation, structure score, and increasingly refined approaches [47]. GB is resistant to high dimensionality and linearity [9].
eXtreme gradient boosting (XGBoost, XG)
XGBoost is a high-performance boosting technique that minimizes the loss function and is optimized by various arrangements. It is a gradient boosting method that loops through loops to add models to a community iteratively. The basic principle behind boosting is to focus on instances that the model cannot predict correctly or that are difficult. These instances are given more emphasis by skewing the distribution of observations to make such measures appear in a sample probable. Therefore, the next weak student will focus more on guessing hard instances correctly. By combining all the simple prediction rules into one overarching model, a powerful predictor, XGBoost, is obtained. [37].
2.2.3 Feature subset selection
This method is used to select the most relevant features of all features during model building. It reduces the complexity of the prediction model [19]. In the study, I applied feature selection with random forest in the Python programming language to construct a model that only contains the most significant features. Classification input parameters were obtained using this algorithm.
Two selection strategies were used, MLR followed by random forest feature subset selection, to: (i) reduce the number of features fed to classifiers, (ii) find the most relevant and uncorrelated components, and (iii) reduce computational complexity and prediction time.
2.2.4 Performance evaluation
To evaluate the performance of the classifications used in this study, the dataset was divided into training and test datasets of 75% and %25, respectively. These datasets were randomly separated.
The confusion matrix was used to compare the performance of the classifications. Table 1 showed the four parts of the matrix called True Positive (TP), False Positive (FP), True Negative (TN), and False Negative (FN).
Non-diabetes and diabetes labeled samples have been considered a positive class and negative class, respectively. Here, TP is the samples of non-diabetes correctly classified as “non-diabetes”, FP is the samples of “diabetes” classified as “non-diabetes “, TN is the samples of diabetes classified as diabetes, and FN is the samples of “non-diabetes” predicted as the “diabetes”. The metrics of precision, recall, F1 score, and accuracy obtained using the confusion matrix were used for performance evaluation, as given in eqs. (1), (2), (3), and (4), respectively.
3 Experimental results and discussions
In the study, classification experiments were performed with hybrid models consisting of testing the validity of variables, dimension reduction, and machine learning methods for the disease diagnosis on early-stage diabetes risk prediction datasets. The multiple linear regression produced output as a numeric vector including the R2 value, the F-statistic and its p value, and an estimate of the error variance, to test the validity of variables. The random forest method was used for dimension reduction; k-NN, gradient boosting, random forest, and XGBoost models were used for classification. Models were tested in the Phyton and Matlab R2021a platforms on a computer with an i5-8250U CPU and 12 GB of RAM.
The research data were formed from 520 instances consisting of 320 instances the “diabetes” and 200 the “non-diabetes”. Figure 2 shows the descriptive statistics and the boxplot of independent variables.
The age range of the participants in the study was 16–90 (mean = 48.03, standard deviation = 12.15). Male participants (328) were more than female participants (192). The least common symptoms in participants were obesity (88 subjects), genital thrush (116), and irritability (126), respectively.
Validity describes the outcome reliability for research that correctly yields the same findings that a researcher wants to assume [39]. Therefore, test reliability is a measure or tool that defines how well it is measured and the accuracy that wants to be seen [5, 31]. The MLR analysis was applied to evaluate the validity of variables. The correlation analysis results of all independent variables with the dependent variable are given in Table 2.
Table 2 shows that the F-static values of the polydipsia and polyuria were 376.4226 and 412.7384, respectively. These values mean that there are significant associations between Diabetes and the two risk factors. After polydipsia and polyuria, gender is the strongest risk factor for diabetes with an F-static value of 130.968. Also, polyuria and polydipsia had the highest R2 (coefficient of determination) among all independent variables.
A p value in linear regression indicates whether the relationship between variables is statistically significant. MLR results showed that the p value for 13 variables was less than 0.05, and these variables were more effective in predicting diabetes. The three predictor variables were not statistically significant because their p values (0.1, 0.28, 0.76) were higher than the usual significance level of 0.05. It could not be used as an input to itching, delayed healing, and obesity prediction models due to the p value.
Dimension reduction methods help to remove irrelevant independent variables from the dataset. With these methods, data storage space gets smaller, and the computational complexity time is also reduced. Thus, the time taken to reach the same goal is shortened. The downside is that they can rarely cause significant data loss [34, 36]. Classification models need to use the most relevant variables instead of unnecessary arguments in their inputs to increase training efficiency. Here, feature selection is performed using the random forest algorithm. This algorithm finds the weights of attributes. Figure 3 shows all the features of this study’s data set.
The random forest algorithm identified the most significant attributes required for the early-stage diabetes risk prediction model. By ranking features, the size of the feature set has been reduced. The seven variables, including delayed healing, visual blurring, weakness, itching, obesity, genital thrush, and muscle stiffness were not used as inputs to the classification models. Because their p-values were equal to or greater than 0.05, or their feature ranking score was low. According to MLR and RF analysis results, the most significant risk factors for diabetes were found polydipsia, polyuria, and gender. Nine features obtained from 16 independent variables were finally used as inputs to classification models for Model II.
Among the 520 samples, 390 samples were randomly selected for training, and the remaining 130 samples were used for testing. All the four classifiers (k-NN, random forest, gradient boosting, XGBoost) were trained with all independent variables for the first experiments (Model I). After the classifier training, the confusion matrices were obtained for the test data, showing the classifiers’ performance. Figure 4 shows the confusion matrices obtained from all the models for the testing phase.
It is found that the RF and XGBoost models outperform the competitive models as it has better and more consistent true positive (51) and true negative (78) values as compared with the other two models. Also, it shows that the RF and XGBoost have lesser false-positive (1) and false-negative (0) values. The k-NN model showed the lowest performance, with false positives of 11 and a false-negative of 7 values.
Results were visualized in confusion matrices. The same matrices were obtained for the RF and XG algorithm. Evaluation metrics were calculated using confusion matrices and presented in Table 3 below.
The macro and weighted averages of the confusion matrix for each classifier were calculated for precision, recall, and F1 score. RF and XG algorithms showed the best performance for all variables. The results show that the RF and XG techniques outperform others in all three categories by achieving 0.99, 0.99, and 0.99% of macro-average precision, recall, and F-measures respectively. The same success was obtained for the weighted average values.
As shown in Table 3, RF and XG algorithms show better results with an accuracy of 99.23%. It gives the highest accuracy as compared to GB and k-NN. However, k-NN showed the lowest accuracy in all four metrics, including accuracy, precision, recall, and F1-score compared to the other three algorithms.
All classifiers were trained with nine independent variables for Model II. After training the models, the confusion matrices needed to measure the model performance of test data were obtained. Figure 5 shows the confusion matrices from all models with nine variables for test data.
It is found that the RF and XGBoost models outperform the competitive models as it has better and more consistent true positive (51) and true negative (78) values as compared with the other two models. Also, it shows that the RF and XGBoost have lesser false-positive (1) and false-negative (0) values. The k-NN model showed the lowest performance, with false positives of 11 and a false-negative of 6 values.
Although the number of variables was reduced, the RF and XG algorithm matrices did not change compared to the first experiment. But the confusion matrix of the k-NN and GB algorithm has gotten better. Evaluation metrics were shown in Table 4 below.
In Model II, unlike the first one, only nine variables were applied to the input of the classifiers. Classifier performances, including recall, precision, F1 score, and accuracy, increased in k-NN and GB, but no change was observed in RF and XG. But still, the XG and RF had better results than the others, with an accuracy of 99.23%. When the proposed feature selection was applied the XG algorithm achieved the best results for all three measures. The best results achieved for macro-average precision, recall, and F-measurements are 0.99, 0.99, and 0.99 respectively when the feature selection algorithm is applied.
The memory usage and computational time decreased for four classification models. Computation times were 0.0245, 0.07563, 0.14635, 0.04825 s for the k-NN, GB, RF and XG models, respectively. Feature selection reduced the computation time from 0.00798 seconds to 0.00245 seconds for k-NN (by 69%). The reduction in computation time was more limited in the other three classifiers. Even though the success rates were equal, the calculation time of the RF method was three times that of the XG method.
The receiver operating characteristic (ROC) curve is a two-dimensional plot that illustrates how well a classifier system works. The x- and y-axes are the false positive rate and true positive rate, respectively, for the predictive test. The diagonal line denotes the ROC curve of a random classifier. Each point in the ROC space is a TP/FP data pair for a discrimination cut-off value of the predictive test [57]. As an alternative to ROC curves, precision-recall curves have been often used in machine learning for assessing classification models [13]. The precision-recall curve shows the relationship between precision and recall [26]. Precision measures the number of positive class predictions that belong to the positive class, while recall measures the number of positive class predictions from all positive samples. The Precision-Recall and ROC curves (and AUCROC) for the diabetes prediction using three machine learning algorithms are illustrated in Fig. 6.
The Precision-Recall curve with an AP score of 0.99 obtained with the proposed XG Boost algorithm also indicates an ideal system. The other graph is constructed with “sensitivity” and “1-specificity” on the vertical and horizontal axis (Fig. 6b). In comparing the “non-diabetes” group and the “diabetes” group, the area under the curve (AUC) was 0.87, 0.98, and 0.99 for the k-NN, GB, and XG models, respectively.
Figure 7 reports the classification results for classifiers with/without feature selection to classify diabetes data. The results showed that the feature selection technique could improve the prediction success of the prediction model and shorten the computation time. When the computational time (0.04825 s) and accuracy (99.23%) of the four classifiers were examined together, the XGBoost model was superior to the others.
By analyzing the results, it has been shown that a combination of MLR, RF, and XG has significant effects on diabetes detection based on the direct questionnaire method. The proposed methodology can distinguish diabetic patients from normal individuals with high accuracy. To investigate the effectiveness of the proposed MLR-RF-XG technique, comparisons have been made with some of the approaches reported in the current literature. Table 5 provides the comparative analysis of the mentioned existing methods and the proposed method.
The researchers measured the performance of J48, KNN, SVM, and RF for the prediction of diabetes mellitus [33]. The accuracy obtained by the techniques is 86.46%, 100%, 77.73%, and 100% in J48, KNN, SVM, and RF, respectively. The accuracy achieved by the RF is 85.0% [56]. A genetic algorithm(GA) with a radial basis function neural network (RBFNN) for prediction accuracy achieved 77.4% [43]. The system developed in [59] acquired 80.84%, 78.53%, and 78.41% accuracy for RF, J48, and neural network, respectively. With mRMR implemented, they have found five features that are best for this classification task. For RF, J48, and neural network with mRMR, they obtained 75.08%, 76.13%, and 75.70% accuracy values, respectively. The system proposed in a previous study [55] achieved 95.42% accuracy by using two-stage K-means and logistic regression algorithms. The accuracy obtained by deep learning (DL) is 98.35%, in another study [1]. Tigga and Garg (2020) proposed a system using RF and found an accuracy of 94.10%. Naz and Ahuja (2020) used DL in their system and found an accuracy of 98.07%. Logistic regression obtained an accuracy of 94.59% [18]. The obtained results in a recent study [15] illustrated that the PCA-RF method performed great with an accuracy of 99.2%. Islam et al. (2020) and Gourisaria et al. (2022) evaluated random forest classifiers for the dataset used in this study [15, 29]. They reported that the random forest using 16 features at its entrance reached the best result with an accuracy rate of about 99%. The prediction success of my methodology is better than that achieved by the RF method on the same dataset. Because 99.23% classification accuracy rate was reached with only nine features. In addition, when XGBoost is used as a classifier instead of RF, the calculation time is reduced. By analyzing the information in Table 5, it is clear that the MLR-RF-XG approach can be regarded as an optimal method of data classification when compared to the most recent studies and especially the study using the same dataset.
The main advantages of the proposed method are: reducing the need for blood testing by using direct questionnaires to diagnose early-stage diabetes, low memory and low computation time for computation, and high estimation accuracy, determining which features are more significant for diagnosis, using fewer features than other some studies in the literature.
The disadvantage of this study is that the dataset used contains very little data, especially for the young population. If diabetes cannot be controlled in young people, unwanted diseases may occur in the early period. Therefore, the proposed method should be tested in this age group. The disadvantage of the feature selection method used is that an additional computational cost is involved in the preprocessing stage of the predictive model building process.
4 Hypothesis and limitations
This study is promising and demonstrates that the proposed MLR-RF-XG technique is a powerful method that can assist in the diagnosis of diabetic patients. However, the study’s limitations are as follows: the research sample is limited to only one country and one hospital. The increasing number of instances obtained from different countries and hospitals will further strengthen the confidence in this technique. To further increase the model’s reliability, a plan aiming to increase the number of instances in the existing dataset in the next two years is considered. The second drawback of the system is that the performance of the proposed approach has not been discussed with internists. Therefore, a comparison of this system with internists will be part of a future study.
5 Conclusion
This research proposes a new method for early-stage predict diabetes using XGBoost Classifier with a random forest feature selection technique. The dataset consists of information from questionnaires made to patients of Sylhet Diabetes Hospital in Bangladesh. Analyzes were carried out using different methods to compare the classification performances. The same test/training ratios (25%/75%) were determined for each classifier method used in experimental studies to evaluate the results objectively. The results were compared with the studies in the literature and found superior in terms of classification accuracy, sensitivity, precision, AUC, and F1 score. The proposed method using XGBoost compared to three other classifiers with the same procedures, including k-Nearest Neighbor, random forest, and gradient boosting. Using XGBoost classifier with random forest feature selection technique provided 99.23%, 99%, 99%, 99%, and 0.993 classification accuracy, precision, sensitivity, F1 score, and AUC to identify disease risk factors, respectively. The excellent performance of the XGBoost model shows that it can predict whether early-stage diabetes can be accurate. In this way, the need for blood tests for diagnosis can be reduced. This approach has a practical pre-clinical application in assessing patients at risk for diabetes. The proposed methodology can significantly assist in improving the accuracy of the diagnosis of diabetes and, therefore, for further treatment. Future works will be to test the proposed method with different data sets.
References
Ayon SI, Islam MM (2019) Diabetes prediction: a deep learning approach. Int J Inform Eng Electro Business(IJIEEB) MECS press 11(2):21–27. https://doi.org/10.5815/ijieeb.2019.02.03
Ayon SI, Islam MM, Hossain MR (2020) Coronary artery heart disease prediction: a comparative study of computational intelligence techniques. IETE J Res 2020. https://doi.org/10.1080/03772063.2020.1713916
Bloomgarden ZT (2020) Diabetes and COVID-19. J Diabetes 12:347–348. https://doi.org/10.1111/1753-0407.13027
Breiman L (2001) Random forests. Mach Learn 45:5–32. https://doi.org/10.1023/A:1010933404324
Craig CL, Marshall AL, Sjöström M, Bauman AE, Booth ML, Ainsworth BE, Pratt M, Ekelund U, Yngve A, Sallis JF, Oja P (2003) International physical activity questionnaire: 12-country reliability and validity. Med Sci Sports Exerc 35(8):1381–1395. https://doi.org/10.1249/01.MSS.0000078924.61453.FB
Damle R, Alavi K (2016) The University Healthsystem consortium clinical database: an emerging resource in colorectal surgery research. Sem Colon Rectal Surg 27(2):92–95. https://doi.org/10.1053/j.scrs.2016.01.006
Dong Y, Ma X, Fu T (2021) Electrical load forecasting: a deep learning approach based on K-nearest neighbors. Appl Soft Comput 99:106900. https://doi.org/10.1016/j.asoc.2020.106900
Frank EA, Shubha MC, D’Souza CJM (2012) Blood glucose determination: plasma or serum? J Clin Lab Anal 26(5):317–320. https://doi.org/10.1002/jcla.21524
Friedman JH (2001) Greedy function approximation: a gradient boosting machine. Ann Stat 29(5):1189–1232. https://doi.org/10.1214/aos/1013203451
Garcia-Carretero R, Vigil-Medina L, Mora-Jimenez I, Soguero-Ruiz C, Barquero-Perez O, Ramos-Lopez J (2020) Use of a K-nearest neighbors model to predict the development of type 2 diabetes within 2 years in an obese, hypertensive population. Med Biol Eng Comput 58:991–1002. https://doi.org/10.1007/s11517-020-02132-w
Ghasemi J, Saaidpour S, Brown SD (2007) QSPR study for estimation of acidity constants of some aromatic acids derivatives using multiple linear regression (MLR) analysis. J Mol Struct THEOCHEM 805(1–3):27–32. https://doi.org/10.1016/j.theochem.2006.09.026
Ghorbani H (2020) Ill-conditioning in linear regression models and its diagnostics. J Korean Soc Math Educ Ser B: Pure Appl Math 27(2):71–81. https://doi.org/10.7468/jksmeb.2020.27.2.71
Giglioni V, García-Macías E, Venanzi I, Ierimonti L, Ubertini F (2021) The use of receiver operating characteristic curves and precision-versus-recall curves as performance metrics in unsupervised structural damage classification under changing environment. Eng Struct 246(2021):113029. https://doi.org/10.1016/j.engstruct.2021.113029
Goeijenbier M, van Sloten TT, Slobbe L, Mathieu C, van Genderen P, Beyer WEP, Osterhaus ADME (2017) Benefits of flu vaccination for persons with diabetes mellitus: a review. Vaccine 35(38):5095–5101. https://doi.org/10.1016/j.vaccine.2017.07.095
Gourisaria MK, Jee G, Harshvardhan GM, Singh V, Singh PK, Workneh TC (2022) Data science appositeness in diabetes mellitus diagnosis for healthcare systems of developing nations. IET Commun. 16. pp. 532– 547 https://doi.org/10.1049/cmu2.12338
Gündoğdu S (2021) Improving breast cancer prediction using a pattern recognition network with optimal feature subsets. Croatian Med J 62(5):480–487. https://doi.org/10.3325/cmj.2021.62.480
Guo W, Li M, Dong Y, Zhou H, Zhang Z, Tian C, Qin R, Wang H, Shen Y, Du K, Zhao L, Fan H, Luo S, Hu D (2020) Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab Res Rev 36:e3319. https://doi.org/10.1002/dmrr.3319
Gupta D, Choudhury A, Gupta U, Singh P, Prasad M (2021) Computational approach to clinical diagnosis of diabetes disease: a comparative study. Multimed Tools Appl 80:30091–30116. https://doi.org/10.1007/s11042-020-10242-8
Gupta VK, Gupta A, Kumar D, Sardana A (2021) Prediction of COVID-19 confirmed, death, and cured cases in India using random forest model. Big data mining Analy 4(2):116–123. https://doi.org/10.26599/BDMA.2020.9020016
Haque MR, Islam MM, Iqbal H, Reza MS, Hasan MK (2018) Performance evaluation of random forests and artificial neural networks for the classification of liver disorder. 2018 International conference on computer, communication, chemical, material and electronic engineering (IC4ME2). Rajshahi, Bangladesh. pp. 1–5. https://doi.org/10.1109/IC4ME2.2018.8465658
Hasan MK, Islam MM, Hashem MMA (2016) Mathematical model development to detect breast Cancer using multigene genetic programming. 2016 5th International Conference on Informatics, Electronics and Vision (ICIEV), IEEE, Dhaka, Bangladesh. pp 574-57 https://doi.org/10.1109/ICIEV.2016.7760068
He K, Li Y, Zhu J, Liu H, Lee JE, Amos CI, Hyslop T, Jin J, Lin H, Wei Q, Li Y (2016) Component-wise gradient boosting and false discovery control in survival analysis with high-dimensional covariates. Bioinformatics 32(1):50–57. https://doi.org/10.1093/bioinformatics/btv517
Hodgson K, Morris J, Bridson T, Govan B, Rush C, Ketheesan N (2015) Immunological mechanisms contributing to the double burden of diabetes and intracellular bacterial infections. Immunology 144:171–185. https://doi.org/10.1111/imm.12394
Islam MM, Iqbal H, Haque MR, Hasan MK (2017) Prediction of breast cancer using support vector machine and K-Nearest neighbors. 2017 IEEE region 10 humanitarian technology conference. Dhaka, Bangladesh. pp 226–229 https://doi.org/10.1109/R10-HTC.2017.8288944
Islam Z, Islam M, Asraf A (2020) A combined deep CNN-LSTM network for the detection of novel coronavirus (COVID-19) using X-ray images. Inform Med Unlocked 20:100412. https://doi.org/10.1016/j.imu.2020.100412
Islam MM, Haque MR, Iqbal H, Hasan MM, Hasan M, Kabir MN (2020) Breast cancer prediction: a comparative study using machine learning techniques. SN Comput Sci 1:290. https://doi.org/10.1007/s42979-020-00305-w
Islam MM, Islam MZ, Asraf A, Ding W (2020) Diagnosis of COVID-19 from X-rays using combined CNN-RNN architecture with transfer learning. medRxiv. https://doi.org/10.1101/2020.08.24.20181339v1
Islam MM, Karray F, Alhajj R, Zeng J (2020) A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19). https://arxiv.org/abs/2008.04815
Islam MMF, Ferdousi R, Rahman S, Bushra HY (2020) Likelihood prediction of diabetes at early stage using data mining techniques. Adv Intel Syst Comput https://doi.org/10.1007/978-981-13-8798-2_12
Jackins V, Vimal S, Kaliappan M, Lee MY (2021) AI-based smart prediction of clinical disease using random forest classifier and naive Bayes. J Supercomput 77:5198–5219. https://doi.org/10.1007/s11227-020-03481-x
Javed F, Cheema S (2017) Customer Satisfaction and Customer Perceived Value and its Impact on Customer Loyalty: The Mediational Role of Customer Relationship Management. J Int Bank Comm 22(58):1–14
Johnson R, Zhang T (2014) Learning nonlinear functions using regularized greedy forest. IEEE Transac Patt Analy Mach Intel. pp. 942-954 https://doi.org/10.1109/TPAMI.2013.159
Kandhasamy JP, Balamurali S (2015) Performance analysis of classifier models to predict diabetes mellitus. Procedia Comput Sci 47:45–51. https://doi.org/10.1016/j.procs.2015.03.182
Kilicarslan S, Adem K, Celik M (2020) Diagnosis and classification of cancer using hybrid model based on ReliefF and convolutional neural network. Med Hypotheses 137:109577. https://doi.org/10.1016/j.mehy.2020.109577
Knapp S (2013) Diabetes and infection: is there a link? - a mini-review. Gerontology 59:99–104. https://doi.org/10.1159/000345107
Kononenko I (1994) Estimating attributes: analysis and extensions of RELIEF. Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics) 784 LNCS. pp. 171–182 https://doi.org/10.1007/3-540-57868-4_57
Le NQK, Do DT, Nguyen T-T-D, Le QA (2021) A sequence-based prediction of Kruppel-like factors proteins using XGBoost and optimized features. Gene 787:145643. https://doi.org/10.1016/j.gene.2021.145643
Mohapatra S, Satpathy S, Mohanty SN (2022) A comparative knowledge base development for cancerous cell detection based on deep learning and fuzzy computer vision approach. Multimed Tools Appl 81:24799–24814. https://doi.org/10.1007/s11042-022-12824-0
Morse JM, Barrett M, Mayan M, Olson K, Spiers J (2002) Verification strategies for establishing reliability and validity in qualitative research. Int J Qual Methods 1(2):13–22. https://doi.org/10.1177/160940690200100202
Muhammad LJ, Islam MM, Usman SS, Ayon SI (2020) Predictive data mining models for novel coronavirus (COVID-19) infected ‘patients’ recovery. SN Comput Sci 1:206. https://doi.org/10.1007/s42979-020-00216-w
Naz H, Ahuja S (2020) Deep learning approach for diabetes prediction using PIMA Indian dataset. J Diabetes Metab Disord 14 19(1):391–403. https://doi.org/10.1007/s40200-020-00520-5
Pan Y, Pan Z, Wang Y, Wang W (2020) A new fast search algorithm for exact k-nearest neighbors based on optimal triangle-inequality-based check strategy. Knowl-Based Syst 189:105088. https://doi.org/10.1016/j.knosys.2019.105088
Paul S, Choubey DK (2017) GA_RBF NN: a classification system for diabetes. Int J Biomed Eng Technol 23(1):71–93. https://doi.org/10.1504/ijbet.2017.10003045
Rahman MM, Islam MM, Manik MMH, Islam MR, al-Rakhami MS (2021) Machine learning approaches for tackling novel coronavirus (COVID-19) pandemic. SN COMPUT SCI 2:384. https://doi.org/10.1007/s42979-021-00774-7
Saha P, Sadi MS, Islam M (2021) EMCNet: automated COVID-19 diagnosis from X-ray images using convolutional neural network and Ensemble of Machine Learning Classifiers. Inform Med Unlocked, Elsevier 22:100505. https://doi.org/10.1016/j.imu.2020.100505
Santhosh Kumar HS, Karibasappa K (2022) An effective hybrid deep learning with adaptive search and rescue for brain tumor detection. Multimed Tools Appl 81:17669–17701. https://doi.org/10.1007/s11042-022-12474-2
Shrivastav LK, Jha SK (2020) A gradient boosting machine learning approach in modeling the impact of temperature and humidity on the transmission rate of COVID-19 in India. Appl Intell 51:2727–2739. https://doi.org/10.1007/s10489-020-01997-6
Song X, Waitman LR, Yu ASL, Robbins DC, Hu Y, Liu M (2020) Longitudinal risk prediction of chronic kidney disease in diabetic patients using a temporal-enhanced gradient boosting machine: retrospective cohort study. JMIR Med Inform 8(1):e15510. https://doi.org/10.2196/15510
Tigga NP, Garg S (2020) Prediction of type 2 diabetes using machine learning classification methods. Procedia Comput Sci 167:706–716. https://doi.org/10.1016/j.procs.2020.03.336
Torlay L, Perrone-Bertolotti M, Thomas E, Baciu M (2017) Machine learning–XGBoost analysis of language networks to classify patients with epilepsy. Brain Inform 4:159–169. https://doi.org/10.1007/s40708-017-0065-7
UCI Machine learning Repository. Early stage diabetes risk prediction dataset, Downloaded 2021, https://archive.ics.uci.edu/ml/datasets/Early+stage+diabetes+risk+prediction+dataset.
Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, Wang B, Xiang H, Cheng Z, Xiong Y, Zhao Y, Li Y, Wang X, Peng Z (2020) Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA - J Ame Med Asso 323(11):1061–1069. https://doi.org/10.1001/jama.2020.1585
World Health Organization (n.d.) Diabetes. https://www.who.int/health-topics/diabetes#tab=tab_1.
World Health Organization. Definition and diagnosis of diabetes mellitus and intermediate hyperglycemia (2006). https://www.who.int/diabetes/publications/diagnosis_diabetes2006/en/
Wu H, Yang S, Huang Z, He J, Wang X (2018) Type 2 diabetes mellitus prediction model based on data mining. Inform Med Unlocked 10:100–107. https://doi.org/10.1016/j.imu.2017.12.006
Xu W, Zhang J, Zhang Q, Wei X (2017) Risk prediction of type II diabetes based on random forest model. Proceedings of the 3rd IEEE international conference on advances in electrical and electronics, information, communication and bio-informatics, AEEICB 2017. https://doi.org/10.1109/AEEICB.2017.7972337
Yang S, Berdine G (2017) The receiver operating characteristic (ROC) curve. Southwest Respira Crit care Chron 5(19):34–36. https://doi.org/10.12746/swrccc.v5i19.391
Yang JK, Feng Y, Yuan MY, Yuan SY, Fu HJ, Wu BY, Sun GZ, Yang GR, Zhang XL, Wang L, Xu X, Xu XP, Chan JCN (2006) Plasma glucose levels and diabetes are independent predictors for mortality and morbidity in patients with SARS. Diabet Med 23:623–628. https://doi.org/10.1111/j.1464-5491.2006.01861.x
Zou Q, Qu K, Luo Y, Yin D, Ju Y, Tang H (2018) Predicting diabetes mellitus with machine learning techniques. Front Genet 9(515):1–10. https://doi.org/10.3389/fgene.2018.00515
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interests/competing interests
There are no conflicts or competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gündoğdu, S. Efficient prediction of early-stage diabetes using XGBoost classifier with random forest feature selection technique. Multimed Tools Appl 82, 34163–34181 (2023). https://doi.org/10.1007/s11042-023-15165-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-15165-8