Abstract
Background
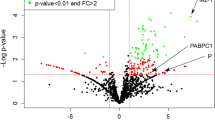
The mitochondrial carrier homolog 2 (MTCH2) is a mitochondrial outer membrane protein regulating mitochondrial metabolism and functions in lipid homeostasis and apoptosis. Experimental data on the interaction of MTCH2 with viral proteins in virus-infected cells are very limited. Here, the interaction of MTCH2 with PA subunit of influenza A virus RdRp and its effects on viral replication was investigated.
Methods
The human MTCH2 protein was identified as the influenza A virus PA-related cellular factor with the Y2H assay. The interaction between GST.MTCH2 and PA protein co-expressed in transfected HEK293 cells was evaluated by GST-pull down. The effect of MTCH2 on virus replication was determined by quantification of viral transcript and/or viral proteins in the cells transfected with MTCH2-encoding plasmid or MTCH2-siRNA. An interaction model of MTCH2 and PA was predicted with protein modeling/docking algorithms.
Results
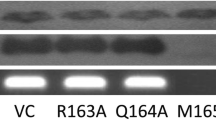
It was observed that PA and GST.MTCH2 proteins expressed in HEK293 cells were co-precipitated by glutathione-agarose beads. The influenza A virus replication was stimulated in HeLa cells whose MTCH2 expression was suppressed with specific siRNA, whereas the increase of MTCH2 in transiently transfected HEK293 cells inhibited viral RdRp activity. The results of a Y2H assay and protein-protein docking analysis suggested that the amino terminal part of the viral PA (nPA) can bind to the cytoplasmic domain comprising amino acid residues 253 to 282 of the MTCH2.
Conclusion
It is suggested that the host mitochondrial MTCH2 protein is probably involved in the interaction with the viral polymerase protein PA to cause negative regulatory effect on influenza A virus replication in infected cells.








Similar content being viewed by others
Data availability
The datasets used during the present study are available from the corresponding author uponreasonable request.
References
Guna A, Stevens TA, Inglis AJ et al (2022) MTCH2 is a mitochondrial outer membrane protein insertase. Science 378(6617):317–322. https://doi.org/10.1126/science.add1856
Palmieri F (2008) Diseases caused by defects of mitochondrial carriers: a review. Biochim Biophys Acta 1777(7–8):564–578. https://doi.org/10.1016/j.bbabio.2008.03.008
Palmieri F (2013) Mitochondrial metabolite carrier protein family. In: Lane MD, Lennarz WJ (eds) Encyclopedia of Biological Chemistry, 2nd edn. Academic, Waltham, pp 149–159
Schwarz M, Andrade-Navarro MA, Gross A (2007) Mitochondrial carriers and pores: key regulators of the mitochondrial apoptotic program? Apoptosis 12(5):869–876. https://doi.org/10.1007/s10495-007-0748-2
Zaltsman Y, Shachnai L, Yivgi-Ohana N et al (2010) MTCH2/MIMP is a major facilitator of tBID recruitment to mitochondria. Nat Cell Biol 12(6):553–562. https://doi.org/10.1038/ncb2057
Maryanovich M, Zaltsman Y, Ruggiero A et al (2015) An MTCH2 pathway repressing mitochondria metabolism regulates haematopoietic stem cell fate. Nat Commun 6:7901. https://doi.org/10.1038/ncomms8901
Rottiers V, Francisco A, Platov M et al (2017) MTCH2 is a conserved regulator of lipid homeostasis. Obes (Silver Spring) 25(3):616–625. https://doi.org/10.1002/oby.21751
Ruggiero A, Aloni E, Korkotian E et al (2017) Loss of forebrain MTCH2 decreases mitochondria motility and calcium handling and impairs hippocampal-dependent cognitive functions. Sci Rep 7:44401. https://doi.org/10.1038/srep44401
Rozenblatt-Rosen O, Deo RC, Padi M et al (2012) Interpreting cancer genomes using systematic host network perturbations by tumour virus proteins. Nature 487(7408):491–495. https://doi.org/10.1038/nature11288
Nainu F, Shiratsuchi A, Nakanishi Y (2017) Induction of apoptosis and subsequent phagocytosis of Virus-infected cells as an antiviral mechanism. Front Immunol 8:1220. https://doi.org/10.3389/fimmu.2017.01220
Rex DAB, Keshava Prasad TS, Kandasamy RK (2022) Revisiting regulated cell death responses in viral infections. Int J Mol Sci 23(13). https://doi.org/10.3390/ijms23137023
Ampomah PB, Lim LHK (2020) Influenza a virus-induced apoptosis and virus propagation. Apoptosis 25(1–2):1–11. https://doi.org/10.1007/s10495-019-01575-3
CaGlayan E, Turan K (2021) Expression profiles of interferon-related genes in cells infected with influenza A viruses or transiently transfected with plasmids encoding viral RNA polymerase. Turk J Biol 45(1):88–103. https://doi.org/10.3906/biy-2005-73
Nayak DP, Hui EK, Barman S (2004) Assembly and budding of influenza virus. Virus Res 106(2):147–165. https://doi.org/10.1016/j.virusres.2004.08.012
Plotch SJ, Bouloy M, Krug RM (1979) Transfer of 5’-terminal cap of globin mRNA to influenza viral complementary RNA during transcription in vitro. Proc Natl Acad Sci U S A 76(4):1618–1622. https://doi.org/10.1073/pnas.76.4.1618
Krug RM, Morgan MA, Shatkin AJ (1976) Influenza viral mRNA contains internal N6-methyladenosine and 5’-terminal 7-methylguanosine in cap structures. J Virol 20(1):45–53. https://doi.org/10.1128/JVI.20.1.45-53.1976
Plotch SJ, Tomasz J, Krug RM (1978) Absence of detectable capping and methylating enzymes in influenza virions. J Virol 28(1):75–83. https://doi.org/10.1128/JVI.28.1.75-83.1978
Shirayama R, Shoji M, Sriwilaijaroen N, Hiramatsu H, Suzuki Y, Kuzuhara T (2016) Inhibition of PA endonuclease activity of influenza virus RNA polymerase by Kampo medicines. Drug Discov Ther 10(2):109–113. https://doi.org/10.5582/ddt.2016.01010
Kim JH, Hatta M, Watanabe S, Neumann G, Watanabe T, Kawaoka Y (2010) Role of host-specific amino acids in the pathogenicity of avian H5N1 influenza viruses in mice. J Gen Virol 91(Pt 5):1284–1289. https://doi.org/10.1099/vir.0.018143-0
Sakabe S, Takano R, Nagamura-Inoue T et al (2013) Differences in cytokine production in human macrophages and in virulence in mice are attributable to the acidic polymerase protein of highly pathogenic influenza a virus subtype H5N1. J Infect Dis 207(2):262–271. https://doi.org/10.1093/infdis/jis523
James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144(4):1425–1436. https://doi.org/10.1093/genetics/144.4.1425
Nagata K, Saito S, Okuwaki M et al (1998) Cellular localization and expression of template-activating factor I in different cell types. Exp Cell Res 240(2):274–281. https://doi.org/10.1006/excr.1997.3930
Pham PTV, Turan K, Nagata K, Kawaguchi A (2018) Biochemical characterization of avian influenza viral polymerase containing PA or PB2 subunit from human influenza a virus. Microbes Infect 20(6):353–359. https://doi.org/10.1016/j.micinf.2018.04.003
Turan K, Mibayashi M, Sugiyama K, Saito S, Numajiri A, Nagata K (2004) Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res 32(2):643–652. https://doi.org/10.1093/nar/gkh192
Sugiyama K, Kawaguchi A, Okuwaki M, Nagata K (2015) pp32 and APRIL are host cell-derived regulators of influenza virus RNA synthesis from cRNA. Elife 4. https://doi.org/10.7554/eLife.08939
Sambrook J, Fritsch ER, Maniatis T (1989) Molecular cloning: a Laboratory Manual. Cold Spring Harbor Laboratory Press. In, Cold Spring Harbor, NY
Gietz RD, Schiestl RH (2007) Large-scale high-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc 2(1):38–41. https://doi.org/10.1038/nprot.2007.15
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. https://doi.org/10.1186/1471-2105-9-40
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596(7873):583–589. https://doi.org/10.1038/s41586-021-03819-2
Varadi M, Anyango S, Deshpande M et al (2022) AlphaFold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50(D1):D439–D444. https://doi.org/10.1093/nar/gkab1061
Kozakov D, Hall DR, Xia B et al (2017) The ClusPro web server for protein-protein docking. Nat Protoc 12(2):255–278. https://doi.org/10.1038/nprot.2016.169
Fan H, Walker AP, Carrique L et al (2019) Structures of influenza a virus RNA polymerase offer insight into viral genome replication. Nature 573(7773):287–290. https://doi.org/10.1038/s41586-019-1530-7
Laskowski RA (2007) Enhancing the functional annotation of PDB structures in PDBsum using key figures extracted from the literature. Bioinformatics 23(14):1824–1827. https://doi.org/10.1093/bioinformatics/btm085
Xue LC, Rodrigues JP, Kastritis PL et al (2016) PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics 32(23):3676–3678. https://doi.org/10.1093/bioinformatics/btw514
Gross A (2005) Mitochondrial carrier homolog 2: a clue to cracking the BCL-2 family riddle? J Bioenerg Biomembr 37(3):113–119. https://doi.org/10.1007/s10863-005-6222-3
Ruprecht JJ, Kunji ERS (2020) The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem Sci 45(3):244–258. https://doi.org/10.1016/j.tibs.2019.11.001
Kodym R, Kodym E, Story MD (2009) 2’-5’-Oligoadenylate synthetase is activated by a specific RNA sequence motif. Biochem Biophys Res Commun 388(2):317–322. https://doi.org/10.1016/j.bbrc.2009.07.167
Chen G, Liu CH, Zhou L, Krug RM (2014) Cellular DDX21 RNA helicase inhibits influenza a virus replication but is counteracted by the viral NS1 protein. Cell Host Microbe 15(4):484–493. https://doi.org/10.1016/j.chom.2014.03.002
Kocmar T, Caglayan E, Rayaman E, Nagata K, Turan K (2022) Human sorting nexin 2 protein interacts with Influenza A virus PA protein and has a negative regulatory effect on the virus replication. Mol Biol Rep 49(1):497–510. https://doi.org/10.1007/s11033-021-06906-9
McKellar J, Rebendenne A, Wencker M, Moncorge O, Goujon C (2021) Mammalian and avian host cell influenza a restriction factors. Viruses 13(3). https://doi.org/10.3390/v13030522
Almond JW (1977) A single gene determines the host range of influenza virus. Nature 270(5638):617–618. https://doi.org/10.1038/270617a0
Subbarao EK, London W, Murphy BR (1993) A single amino acid in the PB2 gene of influenza a virus is a determinant of host range. J Virol 67(4):1761–1764. https://doi.org/10.1128/JVI.67.4.1761-1764.1993
Gao H, Sun H, Hu J et al (2015) Twenty amino acids at the C-terminus of PA-X are associated with increased influenza a virus replication and pathogenicity. J Gen Virol 96(8):2036–2049. https://doi.org/10.1099/vir.0.000143
Gui R, Chen Q (2021) Molecular events involved in Influenza A Virus-Induced cell death. Front Microbiol 12:797789. https://doi.org/10.3389/fmicb.2021.797789
Chen W, Calvo PA, Malide D et al (2001) A novel influenza a virus mitochondrial protein that induces cell death. Nat Med 7(12):1306–1312. https://doi.org/10.1038/nm1201-1306
Ehrhardt C, Wolff T, Pleschka S et al (2007) Influenza a virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J Virol 81(7):3058–3067. https://doi.org/10.1128/JVI.02082-06
Zhirnov OP, Konakova TE, Wolff T, Klenk HD (2002) NS1 protein of influenza a virus down-regulates apoptosis. J Virol 76(4):1617–1625. https://doi.org/10.1128/jvi.76.4.1617-1625.2002
Funding
This work was supported by a grant from the Scientific and Technological Research Council of Turkey (TÜBİTAK; grant no: SBAG-112S518).
Author information
Authors and Affiliations
Contributions
P.U., E.Ç., E.R. and K.T. conducted the experiments. P.U., E.Ç. and K.T. drafted the manuscript. K.N. and K.T. contributed to the conception and design of the study. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
In this study, neither human subjects nor animals were used.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ulupinar, P., Çağlayan, E., Rayaman, E. et al. The mitochondrial carrier homolog 2 is involved in down-regulation of influenza A virus replication. Mol Biol Rep 51, 642 (2024). https://doi.org/10.1007/s11033-024-09584-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09584-5