Abstract
Background
Low-temperature severely limits the growth and development of Camellia oleifera (C. oleifera). The mitogen-activated protein kinase (MAPK) cascade plays a key role in the response to cold stress.
Methods and results
Our study aims to identify MAPK cascade genes in C. oleifera and reveal their roles in response to cold stress. In our study, we systematically identified and analyzed the MAPK cascade gene families of C. oleifera, including their physical and chemical properties, conserved motifs, and multiple sequence alignments. In addition, we characterized the interacting networks of MAPKK kinase (MAPKKK)-MAPK kinase (MAPKK)-MAPK in C. oleifera. The molecular mechanism of cold stress resistance of MAPK cascade genes in wild C. oleifera was analyzed by differential gene expression and real-time quantitative reverse transcription-PCR (qRT-PCR).
Conclusion
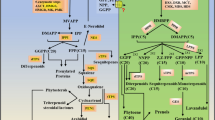
In this study, 21 MAPKs, 4 MAPKKs and 55 MAPKKKs genes were identified in the leaf transcriptome of C. oleifera. According to the phylogenetic results, MAPKs were divided into 4 groups (A, B, C and D), MAPKKs were divided into 3 groups (A, B and D), and MAPKKKs were divided into 2 groups (MEKK and Raf). Motif analysis showed that the motifs in each subfamily were conserved, and most of the motifs in the same subfamily were basically the same. The protein interaction network based on Arabidopsis thaliana (A. thaliana) homologs revealed that MAPK, MAPKK, and MAPKKK genes were widely involved in C. oleifera growth and development and in responses to biotic and abiotic stresses. Gene expression analysis revealed that the CoMAPKKK5/CoMAPKKK43/CoMAPKKK49—CoMAPKK4—CoMAPK8 module may play a key role in the cold stress resistance of wild C. oleifera at a high-elevation site in Lu Mountain (LSG). This study can facilitate the mining and utilization of genetic resources of C. oleifera with low-temperature tolerance.








Similar content being viewed by others
Data availability
The transcriptome data used in this study can be found in online repositories. The name of the repository and accession number can be found below: NCBI Sequence Read Archive, PRJNA915196. In addition to the transcriptome data, all other data generated or analyzed during this study are included in this published article [and its supplementary information files].
References
Qin S, Rong J, Zhang W, Chen J (2018) Cultivation history of Camellia oleifera and genetic resources in the Yangtze River Basin. Biodiv Sci 26(4):384–395. https://doi.org/10.17520/biods.2017254
Peng SF, Lu J, Zhang Z, Ma L, Liu CX, Chen YZ (2020) Global transcriptome and correlation analysis reveal cultivar-specific molecular signatures associated with fruit development and fatty acid determination in Camellia oleifera Abel. Int J Genomics 2020:6162802. https://doi.org/10.1155/2020/6162802
Feng J-L, Yang Z-J, Chen S-P, El-Kassaby YA, Chen H (2017) Signaling pathway in development of Camellia oleifera nurse seedling grafting union. Trees-Struct Funct 31(5):1543–1558. https://doi.org/10.1007/s00468-017-1568-9
Li C, Long Y, Lu M, Zhou J, Wang S, Xu Y, Tan X (2023) Gene coexpression analysis reveals key pathways and hub genes related to late-acting self-incompatibility in Camellia oleifera. Front Plant Sci. https://doi.org/10.3389/fpls.2022.1065872
Lin M, Wang S, Liu Y, Li J, Zhong H, Zou F, Yuan D (2022) Hydrogen cyanamide enhances flowering time in tea oil Camellia (Camellia oleifera Abel.). Ind Crops Prod. https://doi.org/10.1016/j.indcrop.2021.114313
Xie H, Zhang J, Cheng J, Zhao S, Wen Q, Kong P, Zhao Y, Xiang X, Rong J (2023) Field plus lab experiments help identify freezing tolerance and associated genes in subtropical evergreen broadleaf trees: a case study of Camellia oleifera. Front Plant Sci. https://doi.org/10.3389/fpls.2023.1113125
Zhou J, Lu M, Yu S, Liu Y, Yang J, Tan X (2020) In-depth understanding of Camellia oleifera self-incompatibility by comparative transcriptome, proteome and metabolome. Int J Mol Sci. https://doi.org/10.3390/ijms21051600
Knight MR, Knight H (2012) Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol 195(4):737–751. https://doi.org/10.1111/j.1469-8137.2012.04239.x
Wisniewski M, Nassuth A, Teulieres C, Marque C, Rowland J, Cao PB, Brown A (2014) Genomics of cold hardiness in woody plants. Crit Rev Plant Sci 33(2–3):92–124. https://doi.org/10.1080/07352689.2014.870408
Chen J, Yang X, Huang X, Duan S, Long C, Chen J, Rong J (2017) Leaf transcriptome analysis of a subtropical evergreen broadleaf plant, wild oil-tea camellia (Camellia oleifera), revealing candidate genes for cold acclimation. BMC Genom. https://doi.org/10.1186/s12864-017-3570-4
Lin L, Wu J, Jiang M, Wang Y (2021) Plant mitogen-activated protein kinase cascades in environmental stresses. Int J Mol Sci. https://doi.org/10.3390/ijms22041543
Sun TJ, Zhang YL (2022) MAP kinase cascades in plant development and immune signaling. EMBO Rep. https://doi.org/10.15252/embr.202153817
Wang J, Pan C, Wang Y, Ye L, Wu J, Chen L, Zou T, Lu G (2015) Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. https://doi.org/10.1186/s12864-015-1621-2
Colcombet J, Hirt H (2008) Arabidopsis MAPKs: a complex signalling network involved in multiple biological processes. Biochem J 413:217–226. https://doi.org/10.1042/bj20080625
Ichimura K, Shinozaki K, Tena G, Sheen J, Henry Y, Champion A, Kreis M, Zhang SQ, Hirt H, Wilson C, Heberle-Bors E, Ellis BE, Morris PC, Innes RW, Ecker JR, Scheel D, Klessig DF, Machida Y, Mundy J, Ohashi Y, Walker JC, Grp M (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7(7):301–308
Jonak C, Okresz L, Bogre L, Hirt H (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5(5):415–424. https://doi.org/10.1016/s1369-5266(02)00285-6
Hamel L-P, Nicole M-C, Sritubtim S, Morency M-J, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang S, Seguin A, Ellis BE (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11(4):192–198. https://doi.org/10.1016/j.tplants.2006.02.007
Rao KP, Richa T, Kumar K, Raghuram B, Sinha AK (2010) In Silico analysis reveals 75 members of mitogen-activated protein kinase kinase kinase gene family in rice. DNA Res 17(3):139–153. https://doi.org/10.1093/dnares/dsq011
Chatterjee A, Paul A, Unnati GM, Rajput R, Biswas T, Kar T, Basak S, Mishra N, Pandey A, Srivastava AP (2020) MAPK cascade gene family in Camellia sinensis:In-silicoidentification, expression profiles and regulatory network analysis. BMC Genom. https://doi.org/10.1186/s12864-020-07030-x
Paul A, Srivastava AP, Subrahmanya S, Shen GX, Mishra N (2021) In-silico genome wide analysis of mitogen activated protein kinase kinase kinase gene family in C. sinensis. PLoS One. https://doi.org/10.1371/journal.pone.0258657
Kim S-H, Woo D-H, Kim J-M, Lee S-Y, Chung WS, Moon Y-H (2011) Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem Biophys Res Commun 412(1):150–154. https://doi.org/10.1016/j.bbrc.2011.07.064
Teige M, Scheikl E, Eulgem T, Doczi F, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15(1):141–152. https://doi.org/10.1016/j.molcel.2004.06.023
Xing Y, Chen WH, Jia WS, Zhang JH (2015) Mitogen-activated protein kinase kinase 5 (MKK5)-mediated signalling cascade regulates expression of iron superoxide dismutase gene in Arabidopsis under salinity stress. J Exp Bot 66(19):5971–5981. https://doi.org/10.1093/jxb/erv305
Wang C, Lu W, He X, Wang F, Zhou Y, Guo X, Guo X (2016) The cotton mitogen-activated protein kinase kinase 3 functions in drought tolerance by regulating stomatal responses and root growth. Plant Cell Physiol 57(8):1629–1642. https://doi.org/10.1093/pcp/pcw090
Li F, Li M, Wang P, Cox KL Jr, Duan L, Dever JK, Shan L, Li Z, He P (2017) Regulation of cotton (Gossypium hirsutum) drought responses by mitogen-activated protein (MAP) kinase cascade-mediated phosphorylation of GhWRKY59. New Phytol 215(4):1462–1475. https://doi.org/10.1111/nph.14680
Zhao C, Wang P, Si T, Hsu C-C, Wang L, Zayed O, Yu Z, Zhu Y, Dong J, Tao WA, Zhu J-K (2017) MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Dev Cell 43(5):618. https://doi.org/10.1016/j.devcel.2017.09.024
Xie G, Kato F, Imai R (2012) Biochemical identification of the OsMKK6-OsMPK3 signalling pathway for chilling stress tolerance in rice. Biochem J 443:95–102. https://doi.org/10.1042/bj20111792
Kishi-Kaboshi M, Okada K, Kurimoto L, Murakami S, Umezawa T, Shibuya N, Yamane H, Miyao A, Takatsuji H, Takahashi A, Hirochika H (2010) A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J 63(4):599–612. https://doi.org/10.1111/j.1365-313X.2010.04264.x
Eddy SR (2011) Accelerated profile HMM searches. PLoS Comput Biol. https://doi.org/10.1371/journal.pcbi.1002195
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Wang H, Gong M, Guo J, Xin H, Gao Y, Liu C, Dai D, Tang L (2018) Genome-wide identification of Jatropha curcas MAPK, MAPKK, and MAPKKK gene families and their expression profile under cold stress. Sci Rep. https://doi.org/10.1038/s41598-018-34614-1
Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A (2021) Pfam: the protein families database in 2021. Nucleic Acids Res 49(D1):D412–D419. https://doi.org/10.1093/nar/gkaa913
Letunic I, Khedkar S, Bork P (2021) SMART: recent updates, new developments and status in 2020. Nucleic Acids Res 49(D1):D458–D460. https://doi.org/10.1093/nar/gkaa937
Xiao-Lin Z, Bao-Qiang W, Xiao-Hong W (2022) Identification and expression analysis of the CqSnRK2 gene family and a functional study of the CqSnRK2.12 gene in quinoa (Chenopodium quinoa Willd.). BMC Genom 23(1):397. https://doi.org/10.1186/s12864-022-08626-1
Duvaud S, Gabella C, Lisacek F, Stockinger H, Ioannidis V, Durinx C (2021) Expasy, the swiss bioinformatics resource Portal, as designed by its users. Nucleic Acids Res 49(W1):W216–W227. https://doi.org/10.1093/nar/gkab225
Li H, Ran K, Dong Q, Zhao Q, Shi S (2020) Cloning, sequencing, and expression analysis of 32 NAC transcription factors (MdNAC) in apple. PeerJ 8:e8249. https://doi.org/10.7717/peerj.8249
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43(W1):W39–W49. https://doi.org/10.1093/nar/gkv416
Li J, Yu HW, Liu ML, Chen BW, Dong N, Chang XW, Wang JT, Xing SH, Peng HS, Zha LP, Gui SY (2022) Transcriptome-wide identification of WRKY transcription factors and their expression profiles in response to methyl jasmonate in Platycodon grandiflorus. Plant Signaling Behav. https://doi.org/10.1080/15592324.2022.2089473
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. https://doi.org/10.1093/bioinformatics/btp033
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
He Z, Zhang H, Gao S, Lercher MJ, Chen W-H, Hu S (2016) Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res 44(W1):W236–W241. https://doi.org/10.1093/nar/gkw370
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C (2021) The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49(D1):D605–D612. https://doi.org/10.1093/nar/gkaa1074
Danquah A, de Zelicourt A, Boudsocq M, Neubauer J, Frey NFD, Leonhardt N, Pateyron S, Gwinner F, Tamby J-P, Ortiz-Masia D, Marcote MJ, Hirt H, Colcombet J (2015) Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82(2):232–244. https://doi.org/10.1111/tpj.12808
Doczi R, Brader G, Pettko-Szandtner A, Rajh I, Djamei A, Pitzschke A, Teige M, Hirt H (2007) The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 19(10):3266–3279. https://doi.org/10.1105/tpc.106.050039
Takahashi F, Mizoguchi T, Yoshida R, Ichimura K, Shinozaki K (2011) Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol Cell 41(6):649–660. https://doi.org/10.1016/j.molcel.2011.02.029
Takahashi F, Yoshida R, Ichimura K, Mizoguchi T, Seo S, Yonezawa M, Maruyama K, Yamaguchi-Shinozaki K, Shinozaki K (2007) The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 19(3):805–818. https://doi.org/10.1105/tpc.106.046581
Miles GP, Samuel MA, Zhang YL, Ellis BE (2005) RNA interference-based (RNAi) suppression of AtMPK6, an Arabidopsis mitogen-activated protein kinase, results in hypersensitivity to ozone and misregulation of AtMPK3. Environ Pollut 138(2):230–237. https://doi.org/10.1016/j.envpol.2005.04.017
Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415(6875):977–983. https://doi.org/10.1038/415977a
Yoo S-D, Cho Y-H, Tena G, Xiong Y, Sheen J (2008) Dual control of nuclear EIN3 by bifurcate MAPK cascades in C2H4 signalling. Nature 451(7180):789-U781. https://doi.org/10.1038/nature06543
Ren D, Liu Y, Yang K-Y, Han L, Mao G, Glazebrook J, Zhang S (2008) A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA 105(14):5638–5643. https://doi.org/10.1073/pnas.0711301105
Zhou C, Cai Z, Guo Y, Gan S (2009) An Arabidopsis mitogen-activated protein kinase cascade, MKK9-MPK6, plays a role in leaf senescence. Plant Physiol 150(1):167–177. https://doi.org/10.1104/pp.108.133439
Xing Y, Jia W, Zhang J (2008) AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J 54(3):440–451. https://doi.org/10.1111/j.1365-313X.2008.03433.x
Xing Y, Jia W, Zhang J (2009) AtMKK1 and AtMPK6 are involved in abscisic acid and sugar signaling in Arabidopsis seed germination. Plant Mol Biol 70(6):725–736. https://doi.org/10.1007/s11103-009-9503-0
Kong Q, Qu N, Gao MH, Zhang ZB, Ding XJ, Yang F, Li YZ, Dong OX, Chen S, Li X, Zhang YL (2012) The MEKK1-MKK1/MKK2-MPK4 kinase cascade negatively regulates immunity mediated by a mitogen-activated protein kinase kinase kinase in Arabidopsis. Plant Cell 24(5):2225–2236. https://doi.org/10.1105/tpc.112.097253
Minghui G, Jinman L, Dongling B, Zhibin Z, Fang C, Sanfeng C, Yuelin Z (2008) MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res 18(12):1190–1198. https://doi.org/10.1038/cr.2008.300
Zeng Q, Sritubtim S, Ellis BE (2011) AtMKK6 and AtMPK13 are required for lateral root formation in Arabidopsis. Plant Signaling Behav 6(10):1436–1439. https://doi.org/10.4161/psb.6.10.17089
Liu Z, Lv Y, Zhang M, Liu Y, Kong L, Zou M, Lu G, Cao J, Yu X (2013) Identification, expression, and comparative genomic analysis of the IPT and CKX gene families in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. https://doi.org/10.1186/1471-2164-14-594
Hong S, Lim YP, Kwon SY, Shin AY, Kim YM (2021) Genome-wide comparative analysis of flowering-time genes; insights on the gene family expansion and evolutionary perspective. Front Plant Sci. https://doi.org/10.3389/fpls.2021.702243
Tanoue T, Adachi M, Moriguchi T, Nishida E (2000) A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat Cell Biol 2(2):110–116. https://doi.org/10.1038/35000065
Chen L, Hu W, Tan S, Wang M, Ma Z, Zhou S, Deng X, Zhang Y, Huang C, Yang G, He G (2012) Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS One. https://doi.org/10.1371/journal.pone.0046744
Acknowledgements
The authors thank the National Natural Science Foundation of China (32260306) and the National Natural Science Foundation of China (32270238) for funding and supporting of this study.
Funding
This study was funded by the National Natural Science Foundation of China (32260306) and the National Natural Science Foundation of China (32270238).
Author information
Authors and Affiliations
Contributions
K.X., J.Z., X.X., Y.Z. and J.R. designed the experiments. K.X. conducted the experiments with help from J.Z., H.X., L.Z., H.Z., L.F., J.Z., X.X. and Y.Z. K.X., J.Z., H.X., L.Z., H.Z., L.F., J.Z., X.X., Y.Z. and J.R. participated in data analyses. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This declaration does not apply to our study.
Consent to participate
This declaration does not apply to our study.
Consent to publish
This declaration does not apply to our study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2024_9551_MOESM1_ESM.tif
Supplementary file1 (TIF 2345 KB)—Supplementary Fig 1 Gene expression levels of differentially expressed genes (DEGs) through RNA-seq and qRT-PCR analyses. Data are presented as the mean between treated and control samples at each time point ± standard error (SE).
11033_2024_9551_MOESM2_ESM.xlsx
Supplementary file2 (XLSX 49 KB)—Supplementary Table S0. Author names, affiliation and e-mail address of the corresponding author. Supplementary Table S1. MAPK, MAPKK and MAPKKK families in Arabidopsis thaliana, Oryza sativa and Camellia sinensis. Supplementary Table S2. Characteristics of MAPK, MAPKK and MAPKKK gene families in Camellia oleifera. Supplementary Table S3. Motif analysis of MAPK, MAPKK and MAPKKK families in Camellia oleifera. Supplementary Table S4. Identification of homologous proteins in the Camellia oleifera MAPK cascade genes by the BLASTP algorithm. Supplementary Table S5. Expression data of Camellia oleifera MAPK cascade genes under cold stress. Supplementary Table S6. The primer sequences used for qRT-PCR.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xing, K., Zhang, J., Xie, H. et al. Identification and analysis of MAPK cascade gene families of Camellia oleifera and their roles in response to cold stress. Mol Biol Rep 51, 602 (2024). https://doi.org/10.1007/s11033-024-09551-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09551-0