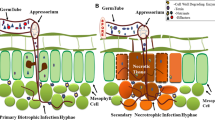
Abstract
Pathogenic fungi and their spores are ubiquitously present and invade the tissues of higher living plants causing pathogenesis and inevitably death or retarded growth. A group of fungi kills its hosts and consume the dead tissues (necrotrophs), while others feed on living tissue (biotrophs) or combination of two (hemibiotrophs). A number of virulent factors is used by fungal pathogens to inhabit new hosts and cause illness. Fungal pathogens develop specialized structures for complete invasion into plant organs to regulate pathogenic growth. Virulence factors like effectors, mycotoxins, cell wall degrading enzymes and organic acids have varied roles depending on the infection strategy and assist the pathogens to possess control on living tissues of the plants. Infection strategies employed by fungi generally masks the plant defense mechanism, however necrotrophs are best known to harm plant tissues with their poisonous secretion. Interestingly, the effector chemicals released by Biotrophs reduce plant cell growth and regulate plant metabolism in their advantage causing no direct death. All these virulence tools cause huge loss to the agricultural product of pre- harvest crops and post-harvest yields causing low output leading to huge economic losses. This review focusses on comprehensive study of range of virulence factors of the pathogenic fungi responsible for their invasion inside the healthy tissues of plants. The compiled information would influence researchers to design antidote against all virulence factors of fungi relevant to their area of research which could pave way for protection against plant pathogenesis.



Similar content being viewed by others
Data availability
There is no new data associated with this article.
References
Naranjo-Ortiz MA, Gabaldón T (2019) Fungal evolution: diversity, taxonomy and phylogeny of the Fungi. Biol Rev Camb Philos Soc 94:2101–2137. https://doi.org/10.1111/brv.12550
Aguilar-Marcelino L, Mendoza-de-Gives P, Al-Ani LKT et al (2020) Chap. 26 - Using molecular techniques applied to beneficial microorganisms as biotechnological tools for controlling agricultural plant pathogens and pest. In: Sharma V, Salwan R, Al-Ani LKTBT-MA of PBM in A (eds). Academic Press, pp 333–349
Doehlemann G, Ökmen B, Zhu W, Sharon A (2017) Plant pathogenic Fungi. https://doi.org/10.1128/microbiolspec.FUNK-0023-2016. Microbiol Spectr 5:
Gao J, Li X, Zhang G et al (2021) Probiotics in the dairy industry—advances and opportunities. Compr Rev Food Sci Food Saf 20. https://doi.org/10.1111/1541-4337.12755
Almeida F, Rodrigues ML, Coelho C (2019) The still underestimated Problem of Fungal diseases Worldwide. Front Microbiol 10:214. https://doi.org/10.3389/fmicb.2019.00214
Kubicek CP, Starr TL, Glass NL (2014) Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu Rev Phytopathol 52:427–451. https://doi.org/10.1146/annurev-phyto-102313-045831
Tan K-C, Oliver RP, Solomon PS, Moffat CS (2010) Proteinaceous necrotrophic effectors in fungal virulence. Funct Plant Biol 37:907–912
González-Fernández R, Prats E, Jorrín-Novo JV (2010) Proteomics of plant pathogenic fungi. J Biomed Biotechnol 2010(932527). https://doi.org/10.1155/2010/932527
Colditz F, Krajinski F, Niehaus K (2007) In: Šamaj J, Thelen JJ (eds) Plant Proteomics upon Fungal Attack BT - Plant Proteomics. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 283–309
Cairney JWG (2005) Basidiomycete mycelia in forest soils: dimensions, dynamics and roles in nutrient distribution. Mycol Res 109:7–20. https://doi.org/10.1017/S0953756204001753
Trail F (2007) Fungal cannons: explosive spore discharge in the Ascomycota. FEMS Microbiol Lett 276:12–18. https://doi.org/10.1111/j.1574-6968.2007.00900.x
Meng S, Torto-Alalibo T, Chibucos MC et al (2009) Common processes in pathogenesis by fungal and oomycete plant pathogens, described with Gene Ontology terms. BMC Microbiol 9:S7. https://doi.org/10.1186/1471-2180-9-S1-S7
Epstein L, Nicholson RL (2006) In: Smith AM, Callow JA (eds) Adhesion and adhesives of Fungi and oomycetes BT - Biological adhesives. Springer Berlin Heidelberg, Berlin, Heidelberg, pp 41–62
Choi W, Dean RA (1997) The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9:1973–1983. https://doi.org/10.1105/tpc.9.11.1973
Du M, Schardl CL, Nuckles EM, Vaillancourt LJ (2005) Using mating-type gene sequences for improved phylogenetic resolution of Collectotrichum species complexes. Mycologia 97:641–658. https://doi.org/10.3852/mycologia.97.3.641
Wiethölter N, Horn S, Reisige K et al (2003) In vitro differentiation of haustorial mother cells of the wheat stem rust fungus, Puccinia graminis f. sp. tritici, triggered by the synergistic action of chemical and physical signals. Fungal Genet Biol 38:320–326. https://doi.org/10.1016/s1087-1845(02)00539-x
Zhao X, Kim Y, Park G, Xu J-R (2005) A mitogen-activated protein kinase cascade regulating infection-related morphogenesis in Magnaporthe Grisea. Plant Cell 17:1317–1329. https://doi.org/10.1105/tpc.104.029116
Lengeler KB, Davidson RC, D’souza C et al (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 64:746–785. https://doi.org/10.1128/MMBR.64.4.746-785.2000
Ligterink W, Kroj T, zur Nieden U et al (1997) Receptor-mediated activation of a MAP kinase in pathogen defense of plants. Science 276:2054–2057. https://doi.org/10.1126/science.276.5321.2054
DeZwaan TM, Carroll AM, Valent B, Sweigard JA (1999) Magnaporthe Grisea Pth11p is a Novel plasma membrane protein that mediates Appressorium differentiation in response to Inductive substrate cues. Plant Cell 11:2013–2030. https://doi.org/10.1105/tpc.11.10.2013
Huang H, Nguyen Thi Thu T, He X et al (2017) Increase of fungal pathogenicity and role of Plant Glutamine in Nitrogen-Induced susceptibility (NIS) to Rice Blast. Front Plant Sci 8. https://doi.org/10.3389/fpls.2017.00265
Cruz-Mireles N, Eseola AB, Osés-Ruiz M et al (2021) From appressorium to transpressorium—defining the morphogenetic basis of host cell invasion by the rice blast fungus. PLoS Pathog 17:e1009779
Wang Z-Y, Jenkinson JM, Holcombe LJ et al (2005) The molecular biology of appressorium turgor generation by the rice blast fungus Magnaporthe Grisea. Biochem Soc Trans 33:384–388. https://doi.org/10.1042/BST0330384
Pei Y, Li X, Zhu Y et al (2019) GhABP19, a Novel Germin-Like protein from Gossypium hirsutum, plays an important role in the regulation of resistance to Verticillium and Fusarium Wilt Pathogens. Front Plant Sci 10. https://doi.org/10.3389/fpls.2019.00583
Liu X-H, Lu J-P, Zhang L et al (2007) Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot Cell 6:997–1005. https://doi.org/10.1128/EC.00011-07
Prats E, Llamas MJ, Jorrin J, Rubiales D (2007) Constitutive coumarin Accumulation on sunflower Leaf Surface prevents Rust Germ Tube Growth and Appressorium differentiation. Crop Sci 47:1119–1124. https://doi.org/10.2135/cropsci2006.07.0482
Bassilana M, Blyth J, Arkowitz RA (2003) Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot Cell 2:9–18. https://doi.org/10.1128/EC.2.1.9-18.2003
Laluk K, Mengiste T (2010) Necrotroph attacks on plants: Wanton Destruction or Covert Extortion? Arabidopsis Book 2010. https://doi.org/10.1199/tab.0136
Gebrie S (2016) Biotrophic Fungi Infection and plant defense mechanism. J Plant Pathol Microbiol 7. https://doi.org/10.4172/2157-7471.1000378
Fones HN, Bebber DP, Chaloner TM et al (2020) Threats to global food security from emerging fungal and oomycete crop pathogens. Nat Food 1:332–342. https://doi.org/10.1038/s43016-020-0075-0
Shao D, Smith DL, Kabbage M, Roth MG (2021) Effectors of Plant Necrotrophic Fungi. Front Plant Sci 12. https://doi.org/10.3389/fpls.2021.687713
Choquer M, Fournier E, Kunz C et al (2007) Botrytis Cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol Lett 277:1–10. https://doi.org/10.1111/j.1574-6968.2007.00930.x
Gupta R, Lee SE, Agrawal GK et al (2015) Understanding the plant-pathogen interactions in the context of proteomics-generated apoplastic proteins inventory. Front Plant Sci 6:352. https://doi.org/10.3389/fpls.2015.00352
Wilson RA, Talbot NJ (2009) Under pressure: investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol 7:185–195. https://doi.org/10.1038/nrmicro2032
Nicaise V, Roux M, Zipfel C (2009) Recent advances in PAMP-Triggered immunity against Bacteria: Pattern Recognition Receptors Watch over and raise the alarm. Plant Physiol 150:1638–1647. https://doi.org/10.1104/pp.109.139709
Zipfel C (2014) Plant pattern-recognition receptors. Trends Immunol 35:345–351. https://doi.org/10.1016/j.it.2014.05.004
Perincherry L, Lalak-Kańczugowska J, Stępień Ł (2019) Fusarium-Produced mycotoxins in Plant-Pathogen interactions. Toxins (Basel) 11. https://doi.org/10.3390/toxins11110664
Pusztahelyi T, Holb I, Pócsi I (2015) Secondary metabolites in fungus-plant interactions. Front Plant Sci 6. https://doi.org/10.3389/fpls.2015.00573
Richard J, Payne G, Desjardins A et al (2003) Mycotoxins: risks in plant, animal and human systems. CAST Task Force Rep 139:101–103
McLean M, Watt MP, Berjak P, Dutton MF (1995) Aflatoxin B1—its effects on an in vitro plant system. Food Addit Contam 12:435–443. https://doi.org/10.1080/02652039509374327
Fadl-Allah EM, Mahmoud MA-H, Abd El-Twab MH, Helmey RK (2011) Aflatoxin B1 induces chromosomal aberrations and 5S rDNA alterations in durum wheat. J Association Arab Universities Basic Appl Sci 10:8–14. https://doi.org/10.1016/j.jaubas.2011.06.002
Hetherington AC, Raistrick H (1931) Studies in the Biochemistry of micro-organisms. Part XIV.—On the production and chemical constitution of a new yellow colouring mater, citrinin, produced from glucose by Penicillium. Philosophical Trans Royal Soc Lond Ser B Containing Papers Biol Character 220:269–295. https://doi.org/10.1098/rstb.1931.0025
Bragulat MR, Martínez E, Castellá G, Cabañes FJ (2008) Ochratoxin A and citrinin producing species of the genus Penicillium from feedstuffs. Int J Food Microbiol 126:43–48. https://doi.org/10.1016/j.ijfoodmicro.2008.04.034
Gelderblom WCA, Thiel PG, Marasas WFO, der Merwe KJ (1984) Natural occurrence of fusarin C, a mutagen produced by Fusarium moniliforme, in corn. J Agric Food Chem 32:1064–1067. https://doi.org/10.1021/jf00125a031
Niehaus E-M, Kleigrewe K, Wiemann P et al (2013) Genetic manipulation of the Fusarium Fujikuroi Fusarin Gene Cluster yields insight into the Complex Regulation and Fusarin Biosynthetic Pathway. Chem Biol 20:1055–1066. https://doi.org/10.1016/j.chembiol.2013.07.004
Arumugam T, Ghazi T, Sheik Abdul N, Chuturgoon AA (2021) Chap. 19 - A review on the oxidative effects of the fusariotoxins: Fumonisin B1 and fusaric acid. In: Patel VB, Preedy VR (eds) Toxicology. Academic Press, pp 181–190
Iqbal N, Czékus Z, Ördög A, Poór P (2023) Fusaric acid-evoked oxidative stress affects plant defence system by inducing biochemical changes at subcellular level. Plant Cell Rep 43:2. https://doi.org/10.1007/s00299-023-03084-9
Ismaiel AA, Papenbrock J (2015) Mycotoxins: producing Fungi and mechanisms of Phytotoxicity. Agriculture 5:492–537. https://doi.org/10.3390/agriculture5030492
Bui-Klimke TR, Wu F (2015) Ochratoxin A and Human Health risk: a review of the evidence. Crit Rev Food Sci Nutr 55:1860–1869. https://doi.org/10.1080/10408398.2012.724480
Ringot D, Chango A, Schneider Y-J, Larondelle Y (2006) Toxicokinetics and toxicodynamics of ochratoxin A, an update. Chem Biol Interact 159:18–46. https://doi.org/10.1016/j.cbi.2005.10.106
Faris JD, Zhang Z, Lu H et al (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proceedings of the National Academy of Sciences 107:13544–13549. https://doi.org/10.1073/pnas.1004090107
Gibson DM, King BC, Hayes ML, Bergstrom GC (2011) Plant pathogens as a source of diverse enzymes for lignocellulose digestion. Curr Opin Microbiol 14:264–270. https://doi.org/10.1016/j.mib.2011.04.002
Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344:1879–1900. https://doi.org/10.1016/j.carres.2009.05.021
Nemri A, Saunders DGO, Anderson C et al (2014) The genome sequence and effector complement of the flax rust pathogen Melampsora Lini. Front Plant Sci 5:98. https://doi.org/10.3389/fpls.2014.00098
Sonah H, Deshmukh RK, Bélanger RR (2016) Computational prediction of Effector proteins in Fungi: opportunities and challenges. Front Plant Sci 7. https://doi.org/10.3389/fpls.2016.00126
Hemetsberger Christoph AND, Herrberger CANDZBANDHMANDDG (2012) The Ustilago maydis Effector Pep1 suppresses Plant immunity by inhibition of host peroxidase activity. PLoS Pathog 8:1–14. https://doi.org/10.1371/journal.ppat.1002684
Hemetsberger C, Mueller AN, Matei A et al (2015) The fungal core effector Pep1 is conserved across smuts of dicots and monocots. New Phytol 206:1116–1126. https://doi.org/10.1111/nph.13304
Misas Villamil JC, Mueller AN, Demir F et al (2019) A fungal substrate mimicking molecule suppresses plant immunity via an inter-kingdom conserved motif. Nat Commun 10:1576. https://doi.org/10.1038/s41467-019-09472-8
Redkar A, Villajuana-Bonequi M, Doehlemann G (2015) Conservation of the Ustilago maydis effector See1 in related smuts. Plant Signal Behav 10:e1086855. https://doi.org/10.1080/15592324.2015.1086855
Tanaka S, Brefort T, Neidig N et al (2014) A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. Elife 3:e01355. https://doi.org/10.7554/eLife.01355
van Esse HP, Bolton MD, Stergiopoulos I et al (2007) The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol Plant-Microbe Interactions® 20:1092–1101. https://doi.org/10.1094/MPMI-20-9-1092
Bolton MD, Van Esse HP, Vossen JH et al (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol 69:119–136. https://doi.org/10.1111/j.1365-2958.2008.06270.x
Kaschani F, Shabab M, Bozkurt T et al (2010) An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol 154:1794–1804. https://doi.org/10.1104/pp.110.158030
Wawra S, Belmonte R, Löbach L et al (2012) Secretion, delivery and function of oomycete effector proteins. Curr Opin Microbiol 15:685–691. https://doi.org/10.1016/j.mib.2012.10.008
Stephenson S-A, Hatfield J, Rusu AG et al (2000) CgDN3: an essential pathogenicity gene of Colletotrichum gloeosporioides necessary to avert a hypersensitive-like response in the host stylosanthes guianensis. Mol Plant-Microbe Interactions® 13:929–941. https://doi.org/10.1094/MPMI.2000.13.9.929
Yoshino K, Irieda H, Sugimoto F et al (2012) Cell death of Nicotiana benthamiana is induced by secreted protein NIS1 of Colletotrichum orbiculare and is suppressed by a homologue of CgDN3. Mol Plant Microbe Interact 25:625–636. https://doi.org/10.1094/MPMI-12-11-0316
Lu X, Miao J, Shen D, Dou D (2022) Proteinaceous Effector Discovery and characterization in Plant Pathogenic Colletotrichum Fungi. Front Microbiol 13. https://doi.org/10.3389/fmicb.2022.914035
Oliver RP, Friesen TL, Faris JD, Solomon PS (2011) Stagonospora nodorum: from Pathology to Genomics and Host Resistance. Annu Rev Phytopathol 50:23–43. https://doi.org/10.1146/annurev-phyto-081211-173019
Li J, Cornelissen B, Rep M (2020) Host-specificity factors in plant pathogenic fungi. Fungal Genet Biol 144:103447. https://doi.org/10.1016/j.fgb.2020.103447
Jiquel A, Gay EJ, Mas J et al (2022) Late effectors from Leptosphaeria maculans as tools for identifying novel sources of resistance in Brassica napus. Plant Direct 6:e435. https://doi.org/10.1002/pld3.435
Jaswal R, Kiran K, Rajarammohan S et al (2020) Effector Biology of Biotrophic Plant Fungal pathogens: current advances and future prospects. Microbiol Res 241:126567. https://doi.org/10.1016/j.micres.2020.126567
Patel ZM, Mahapatra R, Jampala SSM (2020) Chap. 11 - Role of fungal elicitors in plant defense mechanism. In: Sharma V, Salwan R, Al-Ani LKT (eds) Molecular Aspects of Plant Beneficial Microbes in Agriculture. Academic Press, pp 143–158
Davidzon M, Alkan N, Kobiler I, Prusky D (2010) Acidification by gluconic acid of mango fruit tissue during colonization via stem end infection by Phomopsis mangiferae. Postharvest Biol Technol 55:71–77. https://doi.org/10.1016/j.postharvbio.2009.08.009
McCaghey M, Willbur J, Smith DL, Kabbage M (2019) The complexity of the Sclerotinia sclerotiorum pathosystem in soybean: virulence factors, resistance mechanisms, and their exploitation to control Sclerotinia stem rot. Trop Plant Pathol 44:12–22. https://doi.org/10.1007/s40858-018-0259-4
Prusky D, Barad S, Ment D, Bi F (2016) The pH modulation by fungal secreted molecules: a mechanism affecting pathogenicity by postharvest pathogens. Isr J Plant Sci 63:22–30. https://doi.org/10.1080/07929978.2016.1151290
Liang X, Liberti D, Li M et al (2015) Oxaloacetate acetylhydrolase gene mutants of Sclerotinia Sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol Plant Pathol 16:559–571. https://doi.org/10.1111/mpp.12211
Kunz C, Vandelle E, Rolland S et al (2006) Characterization of a new, nonpathogenic mutant of Botrytis cinerea with impaired plant colonization capacity. New Phytol 170:537–550. https://doi.org/10.1111/j.1469-8137.2006.01682.x
Jiao W, Liu X, Li Y et al (2022) Organic acid, a virulence factor for pathogenic fungi, causing postharvest decay in fruits. Mol Plant Pathol 23:304–312. https://doi.org/10.1111/mpp.13159
Prusky D, Lichter A (2008) In: Collinge DB, Munk L, Cooke BM (eds) Mechanisms modulating fungal attack in post-harvest pathogen interactions and their control BT - sustainable disease management in a European context. Springer Netherlands, Dordrecht, pp 281–289
Kabbage M, Williams B, Dickman MB (2013) Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia Sclerotiorum. PLoS Pathog 9:1–12. https://doi.org/10.1371/journal.ppat.1003287
Williams B, Kabbage M, Kim H-J et al (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog 7:e1002107. https://doi.org/10.1371/journal.ppat.1002107
Li H, Zhang Z, He C et al (2016) Comparative proteomics reveals the potential targets of BcNoxR, a putative Regulatory subunit of NADPH oxidase of Botrytis Cinerea. Mol Plant Microbe Interact 29:990–1003. https://doi.org/10.1094/MPMI-11-16-0227-R
Dean R, Van Kan JAL, Pretorius ZA et al (2012) The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol 13:414–430. https://doi.org/10.1111/j.1364-3703.2011.00783.x
Drenth A, Guest DI (2016) Fungal and Oomycete Diseases of Tropical Tree Fruit Crops. Annu Rev Phytopathol 54:373–395. https://doi.org/10.1146/annurev-phyto-080615-095944
Pontes M, de CND, Torres-Rêgo M, de Aquino AKS et al (2021) Harpalyce Brasiliana Benth: a prolific source of bioactive flavonoids with antiophidic potential. Phytochem Lett 41:158–167. https://doi.org/10.1016/j.phytol.2020.09.025
Talhinhas P, Azinheira H, Vieira B et al (2014) Overview of the functional virulent genome of the coffee leaf rust pathogen Hemileia vastatrix with an emphasis on early stages of infection. Front Plant Sci 5. https://doi.org/10.3389/fpls.2014.00088
Avelino J, Cristancho M, Georgiou S et al (2015) The coffee rust crises in Colombia and Central America (2008–2013): impacts, plausible causes and proposed solutions. Food Secur 7:303–321. https://doi.org/10.1007/s12571-015-0446-9
Callaway E (2016) Devastating wheat fungus appears in Asia for first time. Nature 532:421–422. https://doi.org/10.1038/532421a
Ismaiel AA, Papenbrock J (2015) Mycotoxins: producing Fungi and mechanisms of Phytotoxicity. Agriculture 5:1–46
Mueller AN, Ziemann S, Treitschke S et al (2013) Compatibility in the Ustilago maydis–maize Interaction requires inhibition of host cysteine proteases by the Fungal Effector Pit2. PLoS Pathog 9:e1003177
Marshall R, Kombrink A, Motteram J et al (2011) Analysis of two in Planta expressed LysM Effector Homologs from the Fungus Mycosphaerella Graminicola reveals Novel Functional properties and varying contributions to virulence on wheat. Plant Physiol 156:756–769. https://doi.org/10.1104/pp.111.176347
de Jonge R, van Esse HP, Kombrink A et al (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329:953–955. https://doi.org/10.1126/science.1190859
Paulus JK, Kourelis J, Ramasubramanian S et al (2020) Extracellular proteolytic cascade in tomato activates immune protease Rcr3. Proceedings of the National Academy of Sciences 117:17409–17417. https://doi.org/10.1073/pnas.1921101117
Song J, Win J, Tian M et al (2009) Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci 106:1654–1659. https://doi.org/10.1073/pnas.0809201106
An B, Wang W, Guo Y et al (2018) BAS2 is required for conidiation and pathogenicity of Colletotrichum gloeosporioides from Hevea brasiliensis. Int J Mol Sci 19. https://doi.org/10.3390/ijms19071860
Bhadauria V, Banniza S, Vandenberg A et al (2013) Overexpression of a novel biotrophy-specific Colletotrichum truncatum effector, CtNUDIX, in hemibiotrophic fungal phytopathogens causes incompatibility with their host plants. Eukaryot Cell 12:2–11. https://doi.org/10.1128/EC.00192-12
Sanz-Martín JM, Pacheco-Arjona JR, Bello-Rico V et al (2016) A highly conserved metalloprotease effector enhances virulence in the maize anthracnose fungus Colletotrichum graminicola. Mol Plant Pathol 17:1048–1062. https://doi.org/10.1111/mpp.12347
Zhu W, Wei W, Fu Y et al (2013) A secretory protein of Necrotrophic Fungus Sclerotinia sclerotiorum that suppresses host resistance. PLoS ONE 8:1–19. https://doi.org/10.1371/journal.pone.0053901
Xiao X, Xie J, Cheng J et al (2014) Novel secretory protein Ss-Caf1 of the plant-pathogenic fungus Sclerotinia Sclerotiorum is required for host penetration and normal sclerotial development. Mol Plant Microbe Interact 27:40–55. https://doi.org/10.1094/MPMI-05-13-0145-R
Lyu X, Shen C, Fu Y et al (2016) A small secreted virulence-related protein is essential for the Necrotrophic Interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog 12:1–28. https://doi.org/10.1371/journal.ppat.1005435
Dallal Bashi Z, Hegedus DD, Buchwaldt L et al (2010) Expression and regulation of Sclerotinia sclerotiorum necrosis and ethylene-inducing peptides (NEPs). Mol Plant Pathol 11:43–53. https://doi.org/10.1111/j.1364-3703.2009.00571.x
Acknowledgements
We would like to acknowledge Amity University, Jharkhand for providing technical support for the preparation of this manuscript.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
KK & RS conceptualized, PK & KK critically reviewed, and PK & RS wrote the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This article did not involve any experiment on human participants or animals.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, P., Sharma, R. & Kumar, K. A perspective on varied fungal virulence factors causing infection in host plants. Mol Biol Rep 51, 392 (2024). https://doi.org/10.1007/s11033-024-09314-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11033-024-09314-x