Abstract
Background
This study aimed to analyze the role of genetic/epigenetic alterations and the prognostic value of the DNAJC9 gene in breast cancer.
Materials and methods
RT-PCR and Q-RT-PCR methods are used to examine DNAJC9 expression in breast cell lines. Survival ratios of breast cancer patients were evaluated by using bc-GenExMiner. Combined bisulfite restriction analysis and UALCAN in-silico tool were used to assess the methylation level of the DNAJC9 promoter. Mutations were searched with the help of Sanger Cosmic database and direct sequencing.
Results
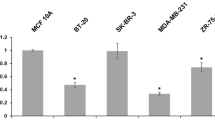
DNAJC9 mRNA expression is significantly higher in basal-like, HER2-Enriched (HER2-E), luminal A and luminal B breast cancer subtypes compared to normal breast-like samples based on DNA microarray datasets (P < 0.001). Similar results were obtained in RNA-seq datasets, except for the luminal A breast cancer subtype (P > 0.1). We did not find any mutation at the core promoter region of DNAJC9 in breast cancer and normal cell lines. Mutations of DNAJC9 are infrequent in clinical samples (<%1). DNAJC9 promoter region is hypomethylated in tumor and normal samples. DNAJC9 expression is unfavorable for survival in basal-like and luminal A breast cancer subtypes.
Conclusions
Mutations or promoter hypomethylation do not appear to have a role in high DNAJC9 gene expression in breast cancer. DNAJC9 expression could be suggested as a novel biomarker in basal-like and luminal A breast cancer subtypes.




Similar content being viewed by others
Data availability
The primer sequences and the datasets used in our study are are provided in online resource file.
References
Sarhadi VK, Armengol G (2022) Molecular biomarkers in Cancer. Biomolecules 12(8):1021. https://doi.org/10.3390/biom12081021
Yun CW, Kim HJ, Lim JH, Lee SH (2019) Heat shock proteins: agents of Cancer Development and therapeutic targets in Anti-Cancer Therapy. Cells 249(1):60. https://doi.org/10.3390/cells9010060
Wu J, Liu T, Rios Z, Mei Q, Lin X, Cao S (2017) Heat shock proteins and Cancer. Trends Pharmacol Sci 38(3):226–256. https://doi.org/10.1016/j.tips.2016.11.009
Lyng H, Brøvig RS, Svendsrud DH et al (2006) Gene expressions and copy numbers associated with metastatic phenotypes of uterine cervical cancer. BMC Genomics. https://doi.org/10.1186/1471-2164-7-268
Van Hoof D, Passier R, Ward-Van Oostwaard D et al (2006) A quest for human and mouse embryonic stem cell-specific proteins. Mol Cell Proteomics 5:1261–1273. https://doi.org/10.1074/mcp.M500405-MCP200
Rappa G, Lorico A (2010) Phenotypic characterization of mammosphereforming cells from the human MA-11 breast carcinoma cell line. Exp Cell Res 316:1576–1586. https://doi.org/10.1016/j.yexcr.2010.01.012
Giotti B, Joshi A, Freeman TC (2017) Meta-analysis reveals conserved cell cycle transcriptional network across multiple human cell types. BMC Genomics 18(1):30. https://doi.org/10.1186/s12864-016-3435-2
Wang C, Zheng W, Yao D et al (2019) Upregulation of DNA metabolism-related genes contributes to Radioresistance of Glioblastoma. Hum Gene Ther Clin Dev 30(2):74–87. https://doi.org/10.1089/humc.2018.251
Zoppino FCM, Guerrero-Gimenez ME, Castro GN, Ciocca DR (2018) Comprehensive transcriptomic analysis of heat shock proteins in the molecular subtypes of human breast cancer. BMC Cancer 18(1):700. https://doi.org/10.1186/s12885-018-4621-1
Buttacavoli M, Di Cara G, D’Amico C et al (2021) Prognostic and functional significant of heat shock proteins (HSPs) in breast Cancer unveiled by multi-omics approaches. Biology (Basel) 10(3):247. https://doi.org/10.3390/biology10030247
Klimczak M, Biecek P, Zylicz A, Zylicz M (2019) Heat shock proteins create a signature to predict the clinical outcome in breast cancer. Sci Rep 9(1):7507. https://doi.org/10.1038/s41598-019-43556-1
Gur-Dedeoglu B, Konu O, Bozkurt B, Ergul G, Seckin S, Yulug IG (2009) Identification of endogenous reference genes for qRT-PCR analysis in normal matched breast tumor tissues. Oncol Res 17(8):353–365. https://doi.org/10.3727/096504009788428460
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Chandrashekar DS, Bashel B, Balasubramanya SAH et al (2017) UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 19(8):649–658. https://doi.org/10.1016/j.neo.2017.05.002 Accessed 14 Mar 2023. http://ualcan.path.uab.edu/index.html
Jézéquel P, Gouraud W, Ben Azzouz F et al (2021) bc-GenExMiner 4.5: new mining module computes breast cancer differential gene expression analyses. Database (Oxford) https://doi.org/10.1093/database/baab007 Accessed 15 Mar 2023. http://bcgenex.ico.unicancer.fr
Jézéquel P, Campone M, Gouraud W et al (2012) bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat 131(3):765–775. https://doi.org/10.1007/s10549-011-1457-7
Parker JS, Mullins M, Cheang MC et al (2009) Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol 27(8):1160–1167. https://doi.org/10.1200/JCO.2008.18.1370
Forbes SA, Beare D, Boutselakis H et al (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45:777–783. https://doi.org/10.1093/nar/gkw1121 Accessed 14 March 2023. https://cancer.sanger.ac.uk/cosmic
Dreos R, Ambrosini G, Périer RC, Bucher P (2015) The Eukaryotic Promoter Database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res 43:92–96. https://doi.org/10.1093/nar/gku1111 Accessed 15 March 2023. https://epd.epfl.ch
Xiong Z, Laird PW (1997) COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res 25(12):2532–2534. https://doi.org/10.1093/nar/25.12.2532
Heiser LM, Sadanandam A, Kuo WL et al (2012) Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA 109:2724–2729. https://doi.org/10.1073/pnas.1018854108
Kent WJ, Sugnet CW, Furey TS et al (2002) The Human Genome Browser at UCSC. Genome Res. 2002;12(6):996–1006. https://doi.org/10.1101/gr.229102 Accessed 14 Mar 2023. http://genome.ucsc.edu
Györffy B, Lanczky A, Eklund AC et al (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 123(3):725–731. https://doi.org/10.1007/s10549-009-0674-9 Accessed 11 Jan 2023. https://kmplot.com
Bouyahya A, Mechchate H, Oumeslakht L et al (2022) The role of epigenetic modifications in human cancers and the Use of Natural Compounds as Epidrugs: mechanistic pathways and pharmacodynamic actions. Biomolecules 12(3):367. https://doi.org/10.3390/biom12030367
Statello L, Guo CJ, Chen LL, Huarte M (2021) Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol 22(2):96–118. https://doi.org/10.1038/s41580-020-00315-9
Cui I, Cui H (2010) Antisense RNAs and epigenetic regulation. Epigenomics 2(1):139–150. https://doi.org/10.2217/epi.09.46
Holliday DL, Speirs V (2011) Choosing the right cell line for breast cancer research. Breast Cancer Res 13(4):215. https://doi.org/10.1186/bcr2889
Chavez KJ, Garimella SV, Lipkowitz S (2011) Triple negative breast cancer cell lines: one tool in the search for better treatment of triple negative breast cancer. Breast Dis 32(1–2):35–48. https://doi.org/10.3233/BD-2010-0307
Zhang Z, Jing J, Ye Y et al (2020) Characterization of the dual functional effects of heat shock proteins (HSPs) in cancer hallmarks to aid development of HSP inhibitors. Genome Med 12(1):101. https://doi.org/10.1186/s13073-020-00795-6
Acknowledgements
We thank Dr. I. Yulug (Bilkent University, Ankara, Turkiye) for providing the cell lines.
Funding
This work was supported by Zonguldak Bulent Ecevit University (Zonguldak, Turkiye) (Grant No: 2020-50737594-02) to Dr. Tolga Acun.
Author information
Authors and Affiliations
Contributions
Conceptualization: TA; Methodology: OI, TA; Formal analysis and investigation: OI, TA; Writing - original draft preparation: TA; Funding acquisition: TA; Supervision: TA.
Corresponding author
Ethics declarations
Competing interests
The authors have no conflicts of interest relevant to this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Incekara, O., Acun, T. DNAJC9 expression in basal-like and luminal A breast cancer subtypes predicts worse survival. Mol Biol Rep 50, 7275–7282 (2023). https://doi.org/10.1007/s11033-023-08654-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08654-4