Abstract
Background
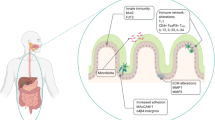
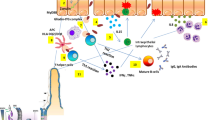
Celiac disease (CD) is a chronic immune-mediated enteropathy and a cytokine network is involved in its pathogenesis. Interleukin-2 (IL-2) has a key role in the adaptive immune pathogenesis of CD and has been reported to be one of the earliest cytokines to be elicited after gluten exposure by CD patients. This study aimed at investigating the expression level of IL-2 and functionally related genes SOCS1 and TBX21 in active and treated CD patients compared to controls.
Methods and results
Peripheral blood (PB) samples were collected from 40 active CD (ACD), 100 treated CD, and 100 healthy subjects. RNA was extracted, cDNA was synthesized and mRNA expression levels of the desired genes were investigated by Real-time PCR. The gene–gene interaction network was also constructed by GeneMANIA. Our results showed a higher PB mRNA expression of IL-2 in ACD patients compared to controls (p = 0.001) and treated CD patients (p˂0.0001). The mRNA expression level of TBX21 was also significantly up-regulated in ACD patients compared to controls (P = 0.03). SOCS1 mRNA level did not differ between active and treated CD patients and controls (p˃0.05) but showed a significant correlation with the patient’s aphthous stomatitis symptom (r = 0.37, p = 0.01). ROC curve analysis suggested that the use of IL-2 levels can reach a high specificity and sensitivity in discriminating active CD patients.
Conclusions
The PB level of IL-2 has the potential to be introduced as a diagnostic biomarker for CD. Larger cohort studies, including pediatric patients, are needed to achieve more insights in this regard.




Similar content being viewed by others
Abbreviations
- ACD:
-
Active celiac disease
- B2M:
-
Beta-2-microglobulin
- CD:
-
Celiac disease
- cDNA:
-
Complementary DNA
- GGI:
-
Gene-gene interaction
- HLA:
-
Human leukocyte antigens
- IFN-γ:
-
interferon gamma
- IL:
-
Interleukin
- JAK:
-
Janus kinase
- NCBI:
-
National Biotechnology Information Center
- ONECUT2:
-
one cut domain family member 2
- PB:
-
Peripheral blood
- RNA:
-
Ribonucleic acid
- ROC:
-
Receiver operating characteristic
- SD:
-
Standard deviation
- SOCS1:
-
Suppressors of cytokine signaling 1
- STAT:
-
Signal transducer and activator of transcription
- TBX21:
-
T-box transcription factor
- TFH:
-
T follicular helper
- Th:
-
T helper
- TNF:
-
Tumor necrosis factor
References
Caio G, Volta U, Sapone A et al (2019) Celiac disease: a comprehensive current review. BMC Med 17:142. https://doi.org/10.1186/s12916-019-1380-z
Rostami Nejad M, Rostami K, Emami M, Zali M, Malekzadeh R (2011) Epidemiology of celiac disease in iran: a review. Middle East J Dig Dis 3:5–12
Singh P, Arora A, Strand TA et al (2018) Global prevalence of Celiac Disease: systematic review and Meta-analysis. Clin Gastroenterol Hepatol 16:823–836e822. https://doi.org/10.1016/j.cgh.2017.06.037
Sarno M, Discepolo V, Troncone R, Auricchio R (2015) Risk factors for celiac disease. Ital J Pediatr 41:57. https://doi.org/10.1186/s13052-015-0166-y
Siddiqui K, Uqaili AA, Rafiq M, Bhutto MA (2021) Human leukocyte antigen (HLA)-DQ2 and -DQ8 haplotypes in celiac, celiac with type 1 diabetic, and celiac suspected pediatric cases. Medicine 100:e24954. https://doi.org/10.1097/md.0000000000024954
Rostami-Nejad M, Romanos J, Rostami K et al (2014) Allele and haplotype frequencies for HLA-DQ in iranian celiac disease patients. World J Gastroenterol 20:6302–6308. https://doi.org/10.3748/wjg.v20.i20.6302
Taraghikhah N, Ashtari S, Asri N et al (2020) An updated overview of spectrum of gluten-related disorders: clinical and diagnostic aspects. BMC Gastroenterol 20:258. https://doi.org/10.1186/s12876-020-01390-0
Asri N, Rostami-Nejad M, Rezaei-Tavirani M, Razzaghi M, Asadzadeh-Aghdaei H, Zali MR (2020) Novel therapeutic strategies for Celiac Disease. Middle East J Dig Dis 12:229–237. https://doi.org/10.34172/mejdd.2020.187
Garrote JA, Gómez-González E, Bernardo D, Arranz E, Chirdo F (2008) Celiac disease pathogenesis: the proinflammatory cytokine network. J Pediatr Gastroenterol Nutr 47(Suppl 1):S27–32. https://doi.org/10.1097/MPG.0b013e3181818fb9
Gaffen SL, Liu KD (2004) Overview of interleukin-2 function, production and clinical applications. Cytokine 28:109–123. https://doi.org/10.1016/j.cyto.2004.06.010
Tye-Din JA, Daveson AJM, Ee HC et al (2019) Elevated serum interleukin-2 after gluten correlates with symptoms and is a potential diagnostic biomarker for coeliac disease. Aliment Pharmacol Ther 50:901–910. https://doi.org/10.1111/apt.15477
Malamut G, Cording S, Cerf-Bensussan N (2019) Recent advances in celiac disease and refractory celiac disease. https://doi.org/10.12688/f1000research.18701.1. F1000Research 8
Penedo-Pita M, Peteiro-Cartelle J (1991) Increased serum levels of interleukin-2 and soluble interleukin-2 receptor in celiac disease. J Pediatr Gastroenterol Nutr 12:56–60. https://doi.org/10.1097/00005176-199101000-00012
Hwang ES, Hong JH, Glimcher LH (2005) IL-2 production in developing Th1 cells is regulated by heterodimerization of RelA and T-bet and requires T-bet serine residue 508. J Exp Med 202:1289–1300. https://doi.org/10.1084/jem.20051044
Workman AM, Jacobs AK, Vogel AJ, Condon S, Brown DM (2014) Inflammation enhances IL-2 Driven differentiation of Cytolytic CD4 T cells. PLoS ONE 9:e89010. https://doi.org/10.1371/journal.pone.0089010
Ross SH, Cantrell DA (2018) Signaling and function of Interleukin-2 in T lymphocytes. Annu Rev Immunol 36:411–433. https://doi.org/10.1146/annurev-immunol-042617-053352
Lazarevic V, Glimcher LH, Lord GM (2013) T-bet:a bridge between innate and adaptive immunity. Nat Rev Immunol 13:777–789. https://doi.org/10.1038/nri3536
Liau NPD, Laktyushin A, Lucet IS et al (2018) The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun 9:1558. https://doi.org/10.1038/s41467-018-04013-1
Sporri B, Kovanen PE, Sasaki A, Yoshimura A, Leonard WJ (2001) JAB/SOCS1/SSI-1 is an interleukin-2–induced inhibitor of IL-2 signaling. Blood 97:221–226. https://doi.org/10.1182/blood.V97.1.221
Yu X, Vargas J, Green PHR, Bhagat G (2021) Innate lymphoid cells and Celiac Disease: current perspective. Cell Mol Gastroenterol Hepatol 11:803–814. https://doi.org/10.1016/j.jcmgh.2020.12.002
Li P, Spolski R, Liao W, Leonard WJ (2014) Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol Rev 261:141–156. https://doi.org/10.1111/imr.12199
del Sutra A, Menegatti S, Fuentealba J et al (2021) In vivo genome-wide CRISPR screens identify SOCS1 as a major intrinsic checkpoint of CD4 + Th1 cell response. Sci Immunol 6:eabe8219. https://doi.org/10.1126/sciimmunol.abe8219
Lahat N, Shapiro S, Karban A, Gerstein R, Kinarty A, Lerner A (1999) Cytokine profile in coeliac disease. Scand J Immunol 49:441–446. https://doi.org/10.1046/j.1365-3083.1999.00523.x
Manavalan JS, Hernandez L, Shah JG et al (2010) Serum cytokine elevations in celiac disease: association with disease presentation. Hum Immunol 71:50–57. https://doi.org/10.1016/j.humimm.2009.09.351
Frisullo G, Nociti V, Iorio R et al (2009) T-bet and pSTAT-1 expression in PBMC from coeliac disease patients: new markers of disease activity. Clin Exp Immunol 158:106–114. https://doi.org/10.1111/j.1365-2249.2009.03999.x
Monteleone I, Monteleone G, Del Vecchio Blanco G et al (2004) Regulation of the T helper cell type 1 transcription factor T-bet in coeliac disease mucosa. Gut 53:1090–1095. https://doi.org/10.1136/gut.2003.030551
Lahdenperä A, Ludvigsson J, Fälth-Magnusson K, Högberg L, Vaarala O (2011) The effect of gluten-free diet on Th1-Th2-Th3-associated intestinal immune responses in celiac disease. Scand J Gastroenterol 46:538–549. https://doi.org/10.3109/00365521.2011.551888
Mazzarella G, MacDonald TT, Salvati VM et al (20030 constitutive activation of the signal transducer and activator of transcription pathway in celiac disease lesions.Am J Pathol162:1845–1855. https://doi.org/10.1016/s0002-9440(10)64319-2
Plaza-Izurieta L, Fernandez-Jimenez N, Irastorza I et al (2015) Expression analysis in intestinal mucosa reveals complex relations among genes under the association peaks in celiac disease. Eur J Hum Genet 23:1100–1105. https://doi.org/10.1038/ejhg.2014.244
Aydemir S, Tekin NS, Aktunç E, Numanoğlu G, Ustündağ Y (2004) Celiac disease in patients having recurrent aphthous stomatitis. Turk J Gastroenterol 15:192–195
Hasan A, Patel H, Saleh H, Youngberg G, Litchfield J, Krishnaswamy G (2013) Remission of severe aphthous stomatitis of celiac disease with etanercept. Clin Mol Allergy 11:6. https://doi.org/10.1186/1476-7961-11-6
van Heel DA, Franke L, Hunt KA, et Al (2007) A genome-wide association study for celiac disease identifies risk variants in the region harboring IL2 and IL21. Nat Genet 39:827–829. https://doi.org/10.1038/ng2058
Burchill MA, Yang J, Vang KB, Farrar MA (2007) Interleukin-2 receptor signaling in regulatory T cell development and homeostasis. Immunol Lett 114:1–8. https://doi.org/10.1016/j.imlet.2007.08.005
Dunne MR, Byrne G, Chirdo FG, Feighery C (2020) Coeliac Disease Pathogenesis: the uncertainties of a well-known Immune mediated disorder. https://doi.org/10.3389/fimmu.2020.01374. Front Immunol 11
Banaganapalli B, Mansour H, Mohammed A et al (2020) Exploring celiac disease candidate pathways by global gene expression profiling and gene network cluster analysis. Sci Rep 10:16290. https://doi.org/10.1038/s41598-020-73288-6
Powell MD, Read KA, Sreekumar BK, Jones DM, Oestreich KJ (2019) IL-12 signaling drives the differentiation and function of a TH1-derived TFH1-like cell population. Sci Rep 9:13991. https://doi.org/10.1038/s41598-019-50614-1
Yang Y, Lv X, Zhan L et al (2021) Case Report: IL-21 and Bcl-6 regulate the proliferation and secretion of tfh and tfr cells in the intestinal Germinal Center of patients with inflammatory bowel disease. https://doi.org/10.3389/fphar.2020.587445. Front Pharmacol 11
Iervasi E, Auricchio R, Strangio A, Greco L, Saverino D (2020) Serum IL-21 levels from celiac disease patients correlates with anti-tTG IgA autoantibodies and mucosal damage. Autoimmunity 53:225–230. https://doi.org/10.1080/08916934.2020.1736047
Long D, Chen Y, Wu H, Zhao M, Lu Q (2019) Clinical significance and immunobiology of IL-21 in autoimmunity. J Autoimmun 99:1–14. https://doi.org/10.1016/j.jaut.2019.01.013
Usui T, Preiss JC, Kanno Y et al (2006) T-bet regulates Th1 responses through essential effects on GATA-3 function rather than on IFNG gene acetylation and transcription. J Exp Med 203:755–766. https://doi.org/10.1084/jem.20052165
Yagi R, Zhu J, Paul WE (2011) An updated view on transcription factor GATA3-mediated regulation of Th1 and Th2 cell differentiation. Int Immunol 23:415–420. https://doi.org/10.1093/intimm/dxr029
Hertweck A, Vila de Mucha M, Barber PR et al (2022) The TH1 cell lineage-determining transcription factor T-bet suppresses TH2 gene expression by redistributing GATA3 away from TH2 genes. Nucleic Acids Res 50:4557–4573. https://doi.org/10.1093/nar/gkac258
Yu J, Li D, Jiang H (2020) Emerging role of ONECUT2 in tumors (review). Oncol Lett 20:328. https://doi.org/10.3892/ol.2020.12192
Dusing MR, Maier EA, Aronow BJ, Wiginton DA (2010) Onecut-2 knockout mice fail to thrive during early postnatal period and have altered patterns of gene expression in small intestine. Physiol Genomics 42:115–125. https://doi.org/10.1152/physiolgenomics.00017.2010
Acknowledgements
The authors would like to acknowledge the Gastroenterology and Liver Diseases Research Center of Shahid Beheshti University of Medical Sciences for their support.
Funding
Research reported in this publication was supported by Gastroenterology and Liver Diseases Research Center, Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Author information
Authors and Affiliations
Contributions
Design of experiments by MRN. Analysis of data by NA and FG. First draft of manuscript by FG and EA and subsequent drafting by MRN, MH, AM, NA and HAA. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
The research was authorized by the Ethics Committee of the Research Institute of Gastroenterology and Liver Diseases of Shahid Beheshti University of Medical Sciences (Ethics code: IR.SBMU.RIGLD.REC.1399.035). Written informed consent was also obtained from participants.
Consent to participate
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ganjali, F., Asri, N., Rostami-Nejad, M. et al. Expression analysis of IL-2, TBX21 and SOCS1 in peripheral blood cells of celiac disease patients reveals the diagnostic potential of IL-2. Mol Biol Rep 50, 4841–4849 (2023). https://doi.org/10.1007/s11033-023-08394-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-023-08394-5