Abstract
Background
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has caused millions of infections and deaths worldwide since its discovery in late 2019 in Wuhan, China. The receptor-binding domain (RBD) of the SARS-CoV-2 spike protein binds to the human angiotensin-converting enzyme-2 (ACE2) receptor, a critical component of the renin-angiotensin system (RAS) that initiates the viral transmission. Most of the critical mutations found in SARS-CoV-2 are associated with the RBD of the spike protein. These mutations have the potential to reduce the efficacy of vaccines and neutralizing antibodies.
Methods
In this review, the structural details of ACE2, RBD and their interactions are discussed. In addition, some critical mutations of RBD and their impact on ACE2-RBD interactions are also discussed.
Conclusion
Preventing the interaction between Spike RBD and ACE2 is considered a viable therapeutic strategy since ACE2 binding by RBD is the first step in virus infection. Because the interactions between the two entities are critical for both viral transmission and therapeutic development, it is essential to understand their interactions in detail.
Similar content being viewed by others
Introduction
SARS-CoV-2 has been reported third amongst coronaviruses which caused severe acute respiratory disease followed by SARS-CoV (2003) and MERS-CoV (2012) all through the last two decades. The whole world has been affected by coronavirus disease 2019 (COVID-19) which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), inflicting havoc on medical systems along with costing millions of lives compared to limited minor populations affected by SARS and MERS. The ~30kb genome of SARS-CoV-2 encodes sixteen non-structural proteins (NSP1-NSP16), four structural proteins comprising Nucleocapsid (N), Membrane (M), Envelope (E), and Spike (S) proteins and six other accessory proteins which are encoded by independent open reading frames (ORF- ORF3a, ORF6, ORF7a, ORF7b, ORF8 and ORF10) [1]. To gain access into a cell and initiate infection, the S-protein interacts with the human Angiotensin-converting enzyme 2 (ACE2) receptor [2]. ACE2 was initially identified in 2003 as the receptor for SARS-CoV [3]. Furin, a proteolytic enzyme, cleaves the SARS-CoV-2 spike protein at a unique cleavage site (SPRRAR↓S) at the interface between the S1 and S2 subunits to produce two subunits, S1 and S2 [4,5,6,7]. The S1 subunit comprises of an N-terminal domain (NTD) and the receptor binding domain (RBD) and is accountable for interaction with the host-cell ACE2 for entry. The S2 subunit comprises of the spike trimeric core and aids in membrane fusion. The structural proteins make up the mature virion while the non-structural proteins are crucial for replication and transcription of the viral genome. The entry of the viral particles encompasses its attachment to the host cell membrane and fusion which are facilitated by the S glycoprotein. The homotrimeric S protein is introduced in numerous copies into the cell membrane of the virion providing it a crown-like form. The monomeric form of S protein is a type I membrane protein and contains 66 N-linked glycans per protein trimer and thus it is ascribed to class I viral fusion proteins epitomized by the influenza virus haemagglutinin protein [4, 8, 9]. According to previous reports [1, 4, 10], S proteins of coronaviruses and haemagglutinin share a common structural organization and conformational transition that aids membrane fusion. Viral infectivity, pathogenesis and host range depend on the receptor recognition by coronaviruses. The SARS-CoV-2 spike protein’s RBD region uniquely selects ACE2 as its receptor making it a popular target for vaccines and antiviral drugs [11]. Pathogenic potential of the SARS-CoV-2 genome may alter due to adaptive variants which might escalate the difficulty in drug and vaccine developments. The current vaccines might pose pronounced challenges owing to viral variants [12]. Global efforts are required to track these variants.. Numerous research groups had isolated and sequenced the SARS-CoV-2 and the resultant sequences were deposited in public databases to expedite the tracking of virus evolution [12]. To tackle the SARS-CoV-2 pandemic and as well as possible future related outbreaks it is imperative to understand the structural details of RBD-ACE2 interactions. This review seeks to provide a detailed summary of the structural information.
The structural details of angiotensin-converting enzyme 2 (ACE2)
The 805 amino acid carboxypeptidase, Angiotensin-converting enzyme 2 (ACE2), shares 42% amino acid homology with angiotensin converting enzyme (ACE), a critical component of the renin-angiotensin system (RAS) [13]. The RAS is an endocrine system that regulates hydromineral balance and cardiovascular function. Through a series of proteolytic cleavage events, the effector peptide angiotensin-II (Ang-II), an active end product of RAS, is formed from angiotensinogen. Angiotensinogen is primarily synthesized in the liver and secreted into the circulation, where it is cleaved at the N-terminus by renin to produce the decapeptide angiotensin-I (Ang-I). The N-terminal residues of Ang-I: Phe-8, His-9 and Ile-12 formed 9 H-bonds with renin. The interactions were mapped with the COCOMAPS web server [14] using the crystal structure of human angiotensinogen complexed with renin (PDB ID: 2 X0B) [15] as a template and the H-bond interactions are presented in Table 1. The Ang-I is then processed by the endothelial ACE into an octapeptide Ang-II by the cleavage of the dipeptide, His-Leu, from the C-terminus of angiotensin-I (Fig. 1). The ACE-Ang-II complex is stabilized by 12 H-bonds. Table 1 shows the details of the H-bond interactions based on the crystal structure of the human angiotensin-converting enzyme in complex with angiotensin-II (PDB ID: 4APH) [16]. When angiotensin I (Ang I) is converted to angiotensin II (Ang II), it binds to angiotensin II type I (AT) and type II receptors in the kidney, adrenal cortex, arterioles, and the brain. Ang II stimulates the release of aldosterone in the adrenal cortex, resulting in sodium and water retention [13, 15,16,17]. ACE can cleave a variety of other substrates, including the vasodilator bradykinin and N-acetyl-Ser-Asp-Lys-Pro (Ac-SDKP), a physiological modulator of haematopoiesis.
While ACE2 (EC 3.4.17.23) is homologous to ACE (EC 3.4.15.1) and functions as an important regulator of the renin-angiotensin system (RAS), there are multiple structural and functional differences. The ACE2 gene is located on chromosome Xp22, in contrast to the ACE gene, which is located on chromosome 17. It consists of 18 exons and 20 introns, most of which are similar to the exons found in the ACE gene [13, 18].
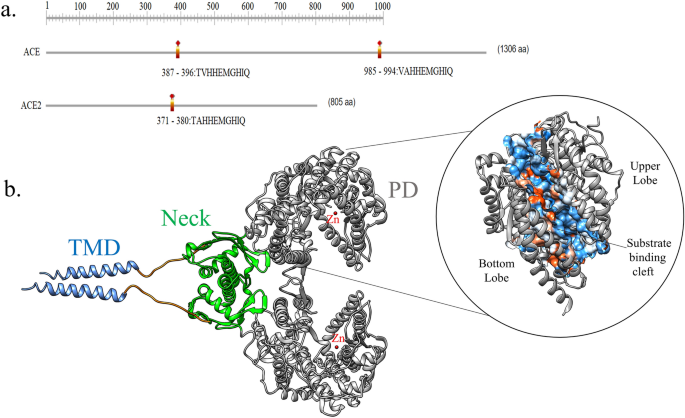
The major structural difference in both zinc metallopeptidases is the active site regions due to which the two enzymes counterbalance rather than reinforce each other’s actions. In contrast to two active sites of ACE (N- and C-terminal domains, each containing a Zn binding motif, HEMGH), ACE2 possesses only a single catalytic domain (Fig. 2a). ACE cleaves C-terminal dipeptide residues from susceptible substrates (a peptidyl dipeptidase), ACE2 acts as a simple carboxypeptidase able to hydrolyze Ang I, forming Ang 1–9 and Ang II to Ang 1–7[18].
The type I membrane protein ACE2 is a homodimer with a claw-shaped extracellular head domain, a small transmembrane domain, and a short intracellular segment (Fig. 2b). The catalytic zinc-binding peptidase domain (PD; residues 19–615) and the smaller neck domain (residues 616–726) compose the head domain, which appears to be where most of the homodimer interactions occur. The PD contains two lobes encompassing the peptide substrate binding site between them. A long linker connects the neck domain to the single-helix transmembrane (TM) domain [20, 21]. The production of Ang 1–9 is processed by the PD which cleaves Ang-I, which is further processed by other enzymes to form Ang 1–7. However, ACE2 can also directly process Ang II to Ang-1-7. The Neck domain is the major contributor to the homodimerization of ACE2, while PD provides a minor interface. The homodimer is stabilized by a network of cation-π interactions and H-bonds. The cation-π interactions are made by Arg-652 and Arg-710 of one protomer (A) with Tyr-641 and Tyr-633 of another protomer (B). The list of H-bond interactions is presented in Table 2. However, no such interactions are observed in PD which depends on non-bonded contacts. Due to the weak interactions of the PDs, they are able to rotate themselves and separate by a distance of ~25 Å while keeping the Neck domain unchanged for the transition from close to open conformation [22].
The structural details of SARS-CoV-2 RBD
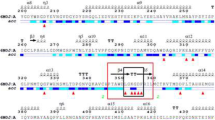
The receptor binding domain (RBD) of both SARS CoV and SARS-CoV-2 uses ACE2 as its receptor. Hence, RBD has become a key therapeutic target. The RBD is a part of the trimeric class I fusion protein Spike (S). To fuse the viral membrane with the host cell membrane, the S-protein undergoes a substantial structural rearrangement from pre-fusion to post-fusion conformations [23]. The 1273 amino acid long S-protein comprises two functional subunits: S1 (residues 14–685), responsible for anchoring to the host cell receptor and S2 (residues 686–1273), responsible for fusion of the viral and cellular membranes (Fig. 3a) [24]. The SARS-CoV-2 RBD (residues 319–541) is present in the S1 subunit, and it appears in two distinct conformations “down” and “up” due to hinge-like conformational movements of the S1 subunit (Fig. 3b). The “down” conformation represents the pre-fusion receptor inaccessible state while the “up” corresponds to the accessible state that is required for ACE2 binding and epitope availability for antibodies [23, 25]. The S1 subunit contains the N-terminal domain (NTD), RBD, sub-domain 1 (SD1) and sub-domain 2 (SD2). There is a high degree of structural homology between individual domains of SARS CoV and SARS-CoV-2 S1 subunit. However, the major difference lies in the RBDs in their respective down conformations: the SARS-CoV RBD in the down conformation packs tightly against the NTD of the protomer partner and is angled closer to the central cavity of the trimer [25]. The crystal structure of SARS-CoV-2 RBD-ACE2 complex (PDB ID: 6M0J) reveals that the RBD contains 7 β strands: β1(residues 354–358), β2 (residues 376–379), β3 (residues 394–403), β4 (residues 432–437), β5 (residues 452–454), β6 (residues 492–494) and β7 (residues 507–516). Out of these, 5 strands namely β1, β2, β3, β4 and β7 are anti-parallel with short connecting helices and loops that form the core [20]. Between the β4 and β7 strands in the core, there is an extended insertion containing the short β5 and β6 strands, α4 and α5 helices and loops. This extended insertion is the receptor-binding motif (RBM) (residues 438–506), which contains most of the contacting residues of SARS-CoV-2 that bind to ACE2.
Details about SARS-CoV-2 RBD-ACE2 interactions
To enter the target cells, the SARS-CoV-2 Spike protein (S) uses human ACE2 as an entry receptor. Hence, the interaction between the SARS-CoV-2 -S and ACE is critical for fusion thereby transmission and pathogenicity of the virus. The importance of this interaction can be inferred from two recent studies which showed that HeLa cells that expressed ACE2 are prone to SARS-CoV-2 infection as opposed to the non ACE2 expressing cells [26] and low nanomolar range (1.2 nM) of binding affinity between RBD and ACE2 [24].The RBM (residues 438–506) of the RBD makes the maximum contact with the N-terminal PD domain of ACE2. A total of 13 H-bonds and 1 salt bridge were observed in the complex (Table 3). The extended RBM contacts the bottom lobe of ACE2 (Fig. 4), where its concave outer surface accommodates the N-terminal helix of the PD [20].
Details about mutational effect on the RBD-ACE2 interactions
Several variants have been reported till date and the structural basis of the effect of the specific mutations on ACE2 binding will be discussed here. Three strains namely, the UK strain with an N501Y mutation in the spike, the South African strain with three concurrent mutations K417N, E484K and N501Y and finally the Brazilian strain with mutations K417T, E484K and N501Y spread rapidly since their first detection [27]. From a structural perspective, N501Y mutation is not expected to contribute significantly towards enhancing complex stabilization since both asparagine and tyrosine can form hydrogen bonds with Tyr-41 of ACE2. Tyrosine substitution could also form pi-pi stacking interaction with Tyr-41 [28]. Indeed, in silico binding energy analysis shows that wild-type spike and N501Y mutant have similar total binding energy values [27, 29]. N501Y might be beneficial for this strain in terms of differential interaction with ACE2 [28]. However, as per earlier in silico reports [27, 29] the African and Brazilian strains of the spike RBD with triple mutations have very strong binding energy interactions with ACE2 as compared to wild type (WT) spike. In the South African variant Asn-417, Lys-484 and Tyr-501 form hydrogen bonds with residues Glu-30, Glu-35 and Glu-75 of ACE2. Lys-484 forms an additional salt bridge with Glu-35 of ACE2. In the case of the Brazilian variant, Glu-35 and Lys-353 of ACE2 formed hydrogen bonds with Lys-484 and Tyr-501 respectively. Salt bridge interaction was observed between Lys-483 and Glu-35. In general, electrostatic interactions were more abundant in both variants as compared to the wild-type spike in complex with ACE2. Similar situation was observed for E484K mutant spike as well. E484K formed H-bonds and salt bridges with Glu-35 of ACE2 which enhanced binding affinity [27].
Two of the most frequently encountered mutations in the SARS-CoV-2 spike are S477N and N439K respectively. In the wild-type spike, Asn-439 forms intra-chain hydrogen bonds with the Spike Ser-443 and Pro-499, thus, not contributing towards ACE2 interaction. However, mutation to the Lys would result in electrostatic interaction with Glu-329 of ACE2 thereby leading to complex stabilization. A similar effect is expected for the S447N mutation which is located in a surface loop of the spike. In the wild-type situation Ser-447 does not engage with ACE2, however, the presence of asparagine would result in the formation of an added H-bond with Ser-19 of ACE2. Both mutations lead to the stabilization of interaction with ACE2 [28].
Spike Q493R is an escape mutation that was first reported in October 2021 [30]. This particular mutation allowed the virus to acquire resistance to Bamlanivimab and Etesevimab which is a monoclonal antibody cocktail therapy [30]. The variant was identified from a patient who contracted COVID and was administered antibody therapy [30]. Gln-493 is an interfacial residue that forms a hydrogen bond with Glu-35 of the ACE2 receptor [28]. Q493R mutation would allow the positively charged arginine side chain to be accommodated within a surface exposed pocket of ACE2 formed by Asp-30, His-34 and Glu-35 respectively [28]. This would result in markedly increased stability of binding. In fact, in silico molecular dynamics simulations demonstrate that the Q493R mutation allows the creation of two extra salt bridges with Glu-35 (more stable) and Asp-38 with ACE2, which are not seen in the wild-type spike.[31]. However, the simulation also indicated the loss of interaction with ACE2 Lys31 and formation of an unfavourable interaction with Lys-353. Q498R similarly enhance binding affinity towards ACE2. In this case, it formed an additional salt bridge with Asp-38. Interactions with Tyr-41 and Gln-42 of ACE2 were stronger compared to WT spike interaction [31].
Spike variants with mutations like N440K and G476S are high affinity ACE2 binders and are located in solvent accessible loop regions. Compared to the WT spike, both mutations lead to drastic conformational alterations wherein, it reorients the RBM loop towards the binding interface thereby allowing additional interactions [31, 32]. Deleterious mutations like G446S and Y505H were also high affinity binders to ACE2 but were found to be destabilizing for RBD [31, 33].
L452R is another notable mutation which interestingly exists in the Delta variant but is absent from the Omicron [34]. Presence of arginine at position 452 results in enhanced infectivity and fusogenic potential [35]. Presence of Leu-452 results in the formation of a hydrophobic patch on the surface of spike RBD due to hydrophobic intramolecular interactions with residues Phe-490 and Leu-492. This patch is non-existent when Leu-452 is replaced by an arginine which affects ACE2 binding resulting in the decreased fusogenic potential of Omicron as compared to the Delta variant [32, 36]. Effects of the RBD mutations on ACE2 binding categorized based on their role are shown in Table 4.
Summary
The rapid outbreak of COVID-19 in Wuhan, China, in December 2019 stunned the world in early 2020. In early March 2020, the situation was designated a pandemic by the World Health Organization (WHO) [37]. Scientists continue to work actively to find new information about the virus that could help combat not only SARS-CoV-2 and its variants, but also future pandemics of this type. Although considerable progress has been made in developing therapeutics against SARS-CoV-2, there are still several obstacles to overcome. One of these problems is that the molecular basis of the interactions of SARS-CoV-2 with the host needs to be further explored. Thus, although a considerable number of host proteins have been reported to interact with SARS-CoV-2 proteins, knowledge of the complex structures of SARS-CoV-2 viral proteins with host proteins is still insufficient [38]. The (S)-glycoprotein of SARS-CoV-2 is the major determinant of host tropism and susceptibility and the main target of antibody responses and the enhanced binding of RBD to the human receptor is one of the critical factors for the transmissibility of the virus. As a result, the appearance of adaptive mutations in the spike protein can have a significant impact on host tropism and viral transmission [39]. Apart from the details of the residues in terms of binding mode, information on how their interaction energies change upon mutation and how these interaction differences affect the fusogenic potential of the virus is critical for the development of future therapeutics.
References
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23(1):3–20
Xia X (2021) Domains and functions of spike protein in SARS-Cov-2 in the context of Vaccine Design. Viruses 13(1):109
Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M (2003) Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426(6965):450–454
Ord M, Faustova I, Loog M (2020) The sequence at spike S1/S2 site enables cleavage by furin and phospho-regulation in SARS-CoV2 but not in SARS-CoV1 or MERS-CoV. Sci Rep 10(1):16944
Jaimes JA, Millet JK, Whittaker GR (2020) Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the Novel S1/S2 site. iScience 23(6):101212
Mjokane N, Maliehe M, Folorunso OS, Ogundeji AO, Gcilitshana OMN, Albertyn J, Pohl CH, Sebolai OM (2022) Cryptococcal protease(s) and the activation of SARS-CoV-2 spike (S) protein. Cells 11(3):437
Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E (2020) The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res 176:104742
Qu Q, Hao P, Xu W, Li L, Jiang Y, Xu Z, Chen J, Gao Z, Pang Z, Jin N, Li C (2022) A vaccine of SARS-CoV-2 S protein RBD induces protective immunity. Int J Mol Sci 23(22):13716
Chawla H, Fadda E, Crispin M (2022) Principles of SARS-CoV-2 glycosylation. Curr Opin Struct Biol 75:102402
Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M (2020) Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369(6501):330–333
Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581(7807):221–224
Han X, Ye Q (2022) The variants of SARS-CoV-2 and the challenges of vaccines. J Med Virol 94(4):1366–1372
Samavati L, Uhal BD (2020) ACE2, much more than just a receptor for SARS-COV-2. Front Cell Infect Microbiol 10:317
Vangone A, Spinelli R, Scarano V, Cavallo L, Oliva R (2011) COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics 27(20):2915–2916
Zhou A, Carrell RW, Murphy MP, Wei Z, Yan Y, Stanley PL, Stein PE, Broughton Pipkin F, Read RJ (2010) A redox switch in angiotensinogen modulates angiotensin release. Nature 468(7320):108–111
Masuyer G, Schwager SL, Sturrock ED, Isaac RE, Acharya KR (2012) Molecular recognition and regulation of human angiotensin-I converting enzyme (ACE) activity by natural inhibitory peptides. Sci Rep 2:717
de Kloet AD, Krause EG, Woods SC (2010) The renin angiotensin system and the metabolic syndrome. Physiol Behav 100(5):525–534
Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, Raizada MK, Grant MB, Oudit GY (2020) Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and Regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res 126(10):1456–1474
Spyroulias GA, Nikolakopoulou P, Tzakos A, Gerothanassis IP, Magafa V, Manessi-Zoupa E, Cordopatis P (2003) Comparison of the solution structures of angiotensin I & II. Implication for structure-function relationship. Eur J Biochem 270(10):2163–2173
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807):215–220
Barros EP, Casalino L, Gaieb Z, Dommer AC, Wang Y, Fallon L, Raguette L, Belfon K, Simmerling C, Amaro RE (2021) The flexibility of ACE2 in the context of SARS-CoV-2 infection. Biophys J 120(6):1072–1084
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367(6485):1444–1448
Zhou T, Tsybovsky Y, Gorman J, Rapp M, Cerutti G, Chuang GY, Katsamba PS, Sampson JM, Schon A, Bimela J, Boyington JC, Nazzari A, Olia AS, Shi W, Sastry M, Stephens T, Stuckey J, Teng IT, Wang P, Wang S, Zhang B, Friesner RA, Ho DD, Mascola JR, Shapiro L, Kwong PD (2020) Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Cell Host Microbe 28(6):867-879e5
Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D (2020) Structure, function, and antigenicity of the SARS-CoV-2 Spike glycoprotein. Cell 181(2):281–292e6
Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS (2020) Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6483):1260–1263
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL (2020) A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273
Khan A, Zia T, Suleman M, Khan T, Ali SS, Abbasi AA, Mohammad A, Wei DQ (2021) Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: an insight from structural data. J Cell Physiol 236(10):7045–7057
Ortuso F, Mercatelli D, Guzzi PH, Giorgi FM (2021) Structural genetics of circulating variants affecting the SARS-CoV-2 spike/human ACE2 complex. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.1886175
Aggarwal A, Naskar S, Maroli N, Gorai B, Dixit NM, Maiti PK (2021) Mechanistic insights into the effects of key mutations on SARS-CoV-2 RBD–ACE2 binding. Phys Chem Chem Phys 23(46):26451–26458
Focosi D, Novazzi F, Genoni A, Dentali F, Gasperina DD, Baj A, Maggi F (2021) Emergence of SARS-COV-2 spike protein escape mutation Q493R after treatment for COVID-19. Emerg Infect Dis 27(10):2728–2731
Jawad B, Adhikari P, Podgornik R, Ching W-Y (2022) Binding interactions between receptor-binding domain of spike protein and human angiotensin converting Enzyme-2 in Omicron variant. J Phys Chem Lett 13(17):3915–3921
Gan HH, Twaddle A, Marchand B, Gunsalus KC (2021) Structural modeling of the SARS-CoV-2 Spike/Human ACE2 Complex Interface can identify High-Affinity Variants Associated with increased transmissibility. J Mol Biol 433(15):167051
Ashoor D, Ben Khalaf N, Marzouq M, Jarjanazi H, Chlif S, Fathallah MD (2021) A computational Approach to evaluate the combined effect of SARS-CoV-2 RBD mutations and ACE2 receptor genetic variants on infectivity: the COVID-19 Host-Pathogen Nexus. Front Cell Infect Microbiol 11:707194
Zhang Y, Zhang T, Fang Y, Liu J, Ye Q, Ding L (2022) SARS-CoV-2 spike L452R mutation increases Omicron variant fusogenicity and infectivity as well as host glycolysis. Signal Transduct Target Ther 7(1):76
Motozono C, Toyoda M, Zahradnik J, Saito A, Nasser H, Tan TS, Ngare I, Kimura I, Uriu K, Kosugi Y, Yue Y, Shimizu R, Ito J, Torii S, Yonekawa A, Shimono N, Nagasaki Y, Minami R, Toya T, Sekiya N, Fukuhara T, Matsuura Y, Schreiber G, Ikeda C, Nakagawa S, Ueno T, Sato K (2021) SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 29(7):1124-1136e11
Cherian S, Potdar V, Jadhav S, Yadav P, Gupta N, Das M, Rakshit P, Singh S, Abraham P, Panda S, N. Team (2021) SARS-CoV-2 spike mutationsin the Second Wave of COVID-19 in Maharashtra India. Microorganisms. 7(1):1542–1214
Cucinotta D, Vanelli M (2020) WHO declares COVID-19 a pandemic. Acta Biomed 91(1):157–160
Yan W, Zheng Y, Zeng X, He B, Cheng W (2022) Structural biology of SARS-CoV-2: open the door for novel therapies. Signal Transduct Target Ther 7(1):26
Escalera A, Gonzalez-Reiche AS, Aslam S, Mena I, Laporte M, Pearl RL, Fossati A, Rathnasinghe R, Alshammary H, van de Guchte A, Farrugia K, Qin Y, Bouhaddou M, Kehrer T, Zuliani-Alvarez L, Meekins DA, Balaraman V, McDowell C, Richt JA, Bajic G, Sordillo EM, Dejosez M, Zwaka TP, Krogan NJ, Simon V, Albrecht RA, van Bakel H, García-Sastre A, Aydillo T (2022) Mutations in SARS-CoV-2 variants of concern link to increased spike cleavage and virus transmission. Cell Host Microbe 30(3):373–387e7
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
SB: conceptualization, writing — original draft & editing; DD & ZH: writing & editing and review.
Corresponding author
Ethics declarations
Competing interest
The authors declare no conflict of interest.
Research involving human and/or animal participants
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Borkotoky, S., Dey, D. & Hazarika, Z. Interactions of angiotensin-converting enzyme-2 (ACE2) and SARS-CoV-2 spike receptor-binding domain (RBD): a structural perspective. Mol Biol Rep 50, 2713–2721 (2023). https://doi.org/10.1007/s11033-022-08193-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08193-4