Abstract
Background
AUX/IAA is an essential signaling molecule and has great physiological importance in various plants, but its function in Zoysia japonica remains unknown.
Methods and results
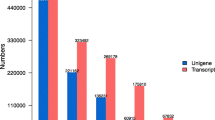
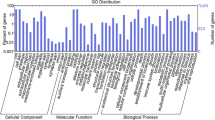
Genome-wide identification and analysis of AUX/IAA genes used bioinformatics methods to investigate the ZjIAA genes' expression of exogenous IAA hydroponics treatment for 2 h by qRT-PCR, control and exogenous IAA treated zoysia were subjected to transcriptome sequencing. ZjIAAs were distributed across the 13 subfamilies by phylogenetic analysis with Oryza sativa and Arabidopsis thaliana. Multiple sequence alignment revealed that the majority of genes were non-canonical ZjIAAs with incomplete domain. The optimal growth concentration of the IAA hormone was 0.05 mM, and the qRT-PCR analysis revealed that eight ZjIAAs were differentially expressed, with seven genes considerably upregulating and one gene significantly downregulating. The result of transcriptome sequencing revealed that 515 differentially expressed genes (DEGs) were identified, with 344 upregulated genes and 171 downregulated genes. A total of 18 genes were annotated as involved in the plant hormone signal transduction pathway. And 8 ZjIAAs exhibited distinct expressions, 7 upregulated, and only one downregulated, according to the qRT-PCR study.
Conclusions
Genome-wide identification and analysis increased the understanding of the evolution and function of the IAA family in zoysia. DEGs of control and treatment with 0.05 mM exogenous IAA hormone were investigated by transcriptome sequencing. ZjIAAs had substantial variations in the expression of associated genes, with the majority of genes upregulated and 18 genes implicated in plant hormone signal transduction.





Similar content being viewed by others
References
Wang L, Zou Y, Kaw HY et al (2020) Recent developments and emerging trends of mass spectrometric methods in plant hormone analysis: a review. Plant Methods 16:1–17. https://doi.org/10.1186/s13007-020-00595-4
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14:290–295. https://doi.org/10.1016/j.pbi.2011.02.001
Tiwari SB, Wang XJ, Hagen G et al (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13:2809–2822. https://doi.org/10.1105/tpc.010289
Boer DR, Freire-Rios A, van den Berg WA et al (2014) Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156:577–589. https://doi.org/10.1016/j.cell.2013.12.027
Szemenyei H, Hannon M, Long JA (2008) TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384–1386. https://doi.org/10.1126/science.1151461
Ramos JA, Zenser N, Leyser O et al (2001) Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13:2349–2360. https://doi.org/10.1105/tpc.010244
Abel S, Oeller PW, Theologis A (1994) Early auxin-induced genes encode short-lived nuclear proteins. PNAS 91:326–330. https://doi.org/10.1073/pnas.91.1.326
Li HT, Wang B, Zhang QH et al (2017) Genome-wide analysis of the auxin/indoleacetic acid (Aux/IAA) gene family in allotetraploid rapeseed (Brassica napus L.). BMC Plant Biol 17:1–14. https://doi.org/10.1186/s12870-017-1165-5
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49:387–400. https://doi.org/10.1023/A:1015255030047
Jain M, Kaur N, Garg R et al (2006) Structure and expression analysis of early auxin-responsive Aux/IAA gene family in rice (Oryza sativa). Funct Integr Genomics 6:47–59. https://doi.org/10.1007/s10142-005-0005-0
Cakir B, Kiliçkaya O, Olcay AC (2013) Genome-wide analysis of Aux/IAA genes in Vitis vinifera: cloning and expression profiling of a grape Aux/IAA gene in response to phytohormone and abiotic stresses. Acta Physiol Plant 35:365–377. https://doi.org/10.1007/s11738-012-1079-7
Mai YX, Wang L, Yang HQ (2011) A gain-of-function mutation in IAA7/AXR2 confers late flowering under short-day light in Arabidopsis F. J Integr Plant Biol 53:480–492. https://doi.org/10.1111/j.1744-7909.2011.01050.x
Jung H, Lee DK, Do Choi Y et al (2015) OsIAA6, a member of the rice Aux/IAA gene family, is involved in drought tolerance and tiller outgrowth. Plant Sci 236:304–312. https://doi.org/10.1016/j.plantsci.2015.04.018
Kloosterman B, Visser RGF, Bachem CWB (2006) Isolation and characterization of a novel potato Auxin/Indole-3-Acetic Acid family member (StIAA2) that is involved in petiole hyponasty and shoot morphogenesis. Plant Physiol Biochem 44:766–775. https://doi.org/10.1016/j.plaphy.2006.10.026
Tian Q, Uhlir NJ, Reed JW (2002) Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. Plant Cell 14:301–319. https://doi.org/10.1105/tpc.010283
Wang L, Feng Z, Wang X et al (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26:136–138. https://doi.org/10.1093/bioinformatics/btp612
Luo H, Sun C, Sun Y et al (2011) Analysis of the transcriptome of Panax notoginseng root uncovers putative triterpene saponin-biosynthetic genes and genetic markers. BMC Genomics 12:1–15. https://doi.org/10.1186/1471-2164-12-S5-S5
Zhang B, Shi JA, Chen JB et al (2016) Efficient virus-induced gene silencing in Cynodon dactylon and Zoysia japonica using rice tungro bacilliform virus vectors. Sci Hortic 207:97–103. https://doi.org/10.1016/j.scienta.2016.05.030
Peterson KW, Fry JD, Bremer DJ (2014) Growth responses of Zoysia spp. under tree shade in the Midwestern United States. HortScience 49:1444–1448. https://doi.org/10.21273/HORTSCI.49.11.1444
Panahi B, Mohammadi SA, Ruzicka K et al (2019) Genome-wide identification and co-expression network analysis of nuclear factor-Y in barley revealed potential functions in salt stress. Physiol Mol Biol Plants 25:485–495. https://doi.org/10.1007/s12298-018-00637-1
Tanaka H, Hirakawa H, Kosugi S (2016) Sequencing and comparative analyses of the genomes of zoysiagrasses. DNA Res 23:171–180. https://doi.org/10.1093/dnares/dsw006
Gasteiger E, Gattiker A, Hoogland C et al (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. https://doi.org/10.1093/nar/gkg563
Garcion C, Béven L, Foissac X (2021) Comparison of current methods for signal peptide prediction in phytoplasmas. Front Microbiol 12:628. https://doi.org/10.3389/fmicb.2021.661524
Krogh A, Larsson B, Von Heijne G et al (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. https://doi.org/10.1006/jmbi.2000.4315
Kohli DK, Bachhawat AK (2003) CLOURE: Clustal Output Reformatter, a program for reformatting ClustalX/ClustalW outputs for SNP analysis and molecular systematics. Nucleic Acids Res 31:3501–3502. https://doi.org/10.1093/nar/gkg502
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Ivica L, Peer B (2007) Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128. https://doi.org/10.1093/bioinformatics/btl529
Hu B, Jin J, Guo A et al (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Bailey TL, Williams N, Misleh C et al (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373. https://doi.org/10.1093/nar/gkl198
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. https://doi.org/10.1093/nar/gkab301
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Bu DC, Luo HT, Huo PP et al (2021) KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res 49:W317–W325. https://doi.org/10.1093/nar/gkab447
Dong D, Zhao Y, Teng K et al (2022) Expression of ZjPSY, a phytoene synthase gene from Zoysia japonica affects plant height and photosynthetic pigment contents. Plants 11:395. https://doi.org/10.3390/plants11030395
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. https://doi.org/10.1126/science.290.5494.1151
Jiang L, Li Z, Yu X et al (2021) Bioinformatics analysis of Aux/IAA gene family in maize. Agron J 113:932–942. https://doi.org/10.1002/agj2.20594
Bonilla AJ, Braun EL, Kimball RT (2010) Comparative molecular evolution and phylogenetic utility of 3′-UTRs and introns in Galliformes. Mol Biol Evol 56:536–542. https://doi.org/10.1016/j.ympev.2010.04.006
Xu G, Guo C, Shan H et al (2012) Divergence of duplicate genes in exon–intron structure. Proc Natl Acad Sci USA 109:1187–1192. https://doi.org/10.1073/pnas.1109047109
Salehin M, Bagchi R, Estelle M (2015) SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27:9–19. https://doi.org/10.1105/tpc.114.133744
Korasick DA, Chatterjee S, Tonelli M et al (2015) Defining a two-pronged structural model for PB1 (Phox/Bem1p) domain interaction in plant auxin responses. J Biol Chem 290:12868–12878. https://doi.org/10.1093/nar/gkg502
Shi Q, Zhang Y, To VT et al (2020) Genome-wide characterization and expression analyses of the auxin/indole-3-acetic acid (Aux/IAA) gene family in barley (Hordeum vulgare L.). Sci Rep 10:1–14. https://doi.org/10.1038/s41598-020-66860-7
Qiao LY, Zhang XJ, Han X et al (2015) A genome-wide analysis of the auxin/indole-3-acetic acid gene family in hexaploid bread wheat (Triticum aestivum L.). Front Plant Sci 6:770. https://doi.org/10.3389/fpls.2015.00770
Gupta R, Dewan I, Bharti R et al (2012) Differential expression analysis for RNA-Seq data. ISRN Bioinf 2012:1–8. https://doi.org/10.5402/2012/817508
Kazan K, Manners JM (2012) JAZ repressors and the orchestration of phytohormone crosstalk. Trends Plant Sci 17:22–31. https://doi.org/10.1016/j.tplants.2011.10.006
Cheng Z, Sun L, Qi T et al (2011) The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 4:279–288. https://doi.org/10.1093/mp/ssq073
Acknowledgements
This research was jointly funded by the Special Fund for Basic Scientific Research Funds of Central Universities (2021ZY84) and the National Natural Science Foundation of China (31971770).
Funding
The work was funded by the Special Fund for Basic Scientific Research Funds of Central Universities (2021ZY84) and the National Natural Science Foundation of China (31971770).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study. Material preparation and data collection were performed by ZY, DD, ZQ and CJ. The first draft of the manuscript and data analysis were performed by ZY and DD. LH and YC selected the topic, modified and finalized the manuscript, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Z., Dong, D., Qi, Z. et al. Genome-wide identification, expression analysis, and transcriptome analysis of the IAA gene family in Zoysia japonica. Mol Biol Rep 50, 4385–4394 (2023). https://doi.org/10.1007/s11033-022-08154-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08154-x