Abstract
Background:
Chickpea (Cicer arietinum L.), a major nutritional source cultivated worldwide, is vulnerable to several abiotic and biotic stresses, including different types of soil-borne pathogens like Fusarium oxysporum f. sp. ciceri, which causes root rot disease and severely affects productivity.
Methods and results
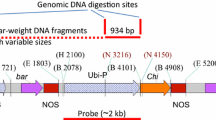
In this study, putative transgenic plants were obtained with the Radish defensin (Rs-AFP2) gene through Agrobacterium tumefaciens mediated transformation using the embryo axis explants. Transgenes were confirmed in 18 putative transgenic plants with PCR-specific primers for nptII and Rs-AFP2 genes. Twelve transgenic plants were established successfully under greenhouse conditions. The T0 plants were allowed for self-pollination to obtain T1 seeds. The T1 plants, selected for Fusarium wilt assay using Fusarium oxysporum f. sp. Cicero, showed different resistance levels, from moderate to high levels in comparison to control plants (wild-type) which exhibited severe wilt symptoms.
Conclusion
Our results suggest the application of Radish defensins (RsAFP1/RsAFP2 genes) for improving pathogen resistance in chickpea.


Similar content being viewed by others
References
Gaur PM, Thudi M, Samineni S (2014) Varshney RK (2014) Advances in chickpea genomics. In: Gupta S, Nadarajan N, Gupta DS (eds) Legumes in Omics era. Springer, New York, pp 73–94
Jacob C, Carrasco B, Schwember AR (2016) Advances in breeding and biotechnology of legume crops. Plant Cell Tissue Organ Cult 127:561–584
Considine MJ, Siddique KHM, Foyer CH (2017) Nature’s pulse power: legumes, food security and climate change. J Expl Bot 68(8):1815–1818
Nene YL, Reddy MV, Haware MP, Ghanekar AM, Amin KS, Pande S, Sharma M (2012) Field diagnostics of chickpea diseases and their control. Information bulletin no. 28 (Revised) International Crops Research Institute for Semi-Arid Tropics (ICRISAT), Patancheru, Hyderabad, India, p 66. ISBN 29-9066-199-2
Leonetti P, Accotto GP, Hanafy MS, Pantaleo V (2018) Viruses and phytoparasitic Nematodes of Cicer arietinum L.: biotechnological approaches in interaction studies and for sustainable control. Front Plant Sci 9:319
Das Bhowmik SS, Cheng AY, Long H, Tan GZH, Hoang TML, Karbaschi MR, Williams B, Higgins TJV, Mundree SG (2019) Robust genetic transformation system to obtain non-chimeric transgenic chickpea. Front Plant Sci 10:524
Pandey A, Yadav R, Kumar S, Kumar A, Shukla P, Sanyal YA (2021) Expression of the entomotoxic Cocculus hirsutus trypsin inhibitor (ChTI) gene in transgenic chickpea enhances its underlying resistance against the infestation of Helicoverpa armigera and Spodoptera litura. Plant Cell Tissue Organ Cult 146:41–56
Das A, Basu PS, Kumar M, Ansari J, Shukla A, Thakur S, Singh P, Datta S, Chaturvedi SK, Sheshshayee MS, Bansal KS, Singh NP (2021) Transgenic chickpea (Cicer arietinum L.) harbouring AtDREB1a are physiologically better adapted to water deficit. BMC Plant Biol 21:39
Thomma BPHJ, Cammue BPA, Thevissen K (2002) Plant defensins. Planta 216:193–202
Feng S, Wang X, Zhang X, Dang PM, Holbrook CC, Culbreath AK, Wu Y, Guo B (2012) Peanut (Arachis hypogaea) expressed sequence tag project: progress and application. Comp Funct Genom 2012:373768
Lacerda AF, Vasconselos EA, Pelegrini PB, Grosside Sa MF (2014) Antifungal defensins and their role in plant defense. Front Microbiol 5:116
Parmar N, Singh KH, Sharma D, Singh L, Kumar P, Nanjundan J, Khan YJ (2017) Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review. 3Biotech 7:239
Campos ML, Liao LM, Alves ESF, Migliolo L, Dias SC, Franco OL (2018) A structural perspective of plant antimicrobial peptides. Biochem J 475:3359–3375
Osborn RW, De Samblanx GW, Thevissen K, Goderis I, Torrekens S, Van Leuven F, Attenborough S, Rees SB, Broekaert WF (1995) Isolation and characterisation of plant defensins from seeds of Asteraceae, Fabaceae, Hippocastanaceae and Saxifragaceae. FEBS Lett 368:257–262
Ceasar AS, Ignacimuthu S (2012) Genetic engineering of crop plants for fungal resistance: role of antifungal genes. Biotechnol Lett 34:995–1002
Moosa A, Farzand A, Sahi ST, Khan SA (2018) Transgenic expression of antifungal pathogenesis-related proteins against phytopathogenic fungi—15 years of success. Isr J Plant Sci 65(1–2):38–54
Ali S, Ganai BA, Kamili AN, Bhat AA, Mir ZA, Bhat JA, Mir ZA, Bhat JA, Grover A (2018) Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol Res 212:29–37
Terras FR, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Vanderleyden J, Cammue BP, Broekaert WF (1995) Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7:573–588
Popov T, Batchvarova R, Slavov S, Docheva P, Alexandrova M, Avetisov V, Serdobinskii L, Korneeva I, Atanassov A (2003) Genetic transformation of tobacco with defensin gene. Biotech Biotech Equip 17(1):17–21
De Bondt A, Zaman S, Broekaert W, Cammue B, Keulemans J (1998) Genetic transformation of apple (Malus pumila Mill.) for increased fungal resistance: in vitro antifungal activity in protein extracts of transgenic apple expressing Rs-AFP2 or Ace-AMP1. Acta Hortic 484:565–570
Parashina EV, Serdobinskii LA, Kalle EG, Lavorova NV, Avetisov VA, Lunin VG, Naroditskii BS (2000) Genetic engineering of oilseed rape and tomato plants expressing a radish defensin gene. Russ J Plant Physiol 47:417–423
Kostov K, Christova P, Slavov S, Batchvarova R (2009) Constitutive expression of a Radish defensin gene Rs-AFP2 in tomato increases the resistance to fungal pathogens. Biotech Biotech Equip 23(1):1121–1125
Jha S, Chattoo BB (2009) Transgene stacking and coordinated expression of plant defensins confer fungal resistance in rice. Rice 2:143–154
Jha S, Chattoo BB (2010) Expression of a plant defensin in rice confers resistance to fungal phytopathogens. Transg Res 19:373–384
Ouyang LJ, He WH, Huang ZC, Zhao LY, Peng SH, Sha YE, Lu ZFH (2012) Introduction of the Rs-AFP2 gene into Eucalyptus urophylla for resistance to Phytophthora capsici. J Trop For Sci 24(2):198–208
Lu Y, Zhang ZY, Ren LJ, Liu BY, Liao Y, Xu HJ, Du LP, Ma HX, Ren ZL, Jing JX, Xin ZY (2009) Molecular analyses on Rs-AFP2 transgenic wheat plants and their resistance to Rhizoctonia cerealis. Acta Agro Sinica 35(4):640–646
Li Z, Zhou M, Zhang Z, Ren L, Du L, Zhang B, Xu H, Xin Z (2011) Expression of a radish defensin in transgenic wheat confers increased resistance to Fusarium graminearum and Rhizoctonia cerealis. Funct Int Gen 11:63–70
Vasavirama K, Kirti PB (2012) Increased resistance to late leaf spot disease in transgenic peanut using a combination of PR genes. Funct Int Gen 12(4):625–634
Vasavirama K, Kirti PB (2013) Tandem combination of Trigonella foenum-graecum defensin (Tfgd2) and Raphanus sativus antifungal protein (RsAFP2) generates a more potent antifungal protein. Funct Int Gen 13(4):435–443
Bala M, Radhakrishnan T, Kumar A, Mishra GP, Dobraia JR, Kirti PB (2016) Overexpression of a fusion defensin gene from radish and fenugreek improvesresistance against leaf spot diseases caused by Cercospora arachidicola and Phaeoisariopsis personata in peanut. Turk J Biol 40(1):139–149
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassay with tobacco tissue cultures. Physiol Plant 15:473–497
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Srivastava J, Sinha P (2016) Direct in vitro shoot regeneration of Cicer arietinum L. from decapitated embryo axes explants. In: Proceedings of international conference on emerging trends engineering & technology, pp 219–222
Sadhu SK, Jogam P, Thampu RK, Abbagani S, Penna S, Peddaboina V (2020) High efficiency plant regeneration and genetic fidelity of regenerants by SCoT and ISSR markers in chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult 141:465–477
Sadhu SK, Jogam P, Gande KK, Banoth R, Penna S, Peddaboina V (2022) Optimization of different factors for an Agrobacterium-mediated genetic transformation system using embryo axis explants of chickpea (Cicer arietinum L.). J Plant Biotechnol 49:61–73. https://doi.org/10.5010/JPB.2022.49.1.061
Jefferson RA, Kavanagh TA, Bevan NW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:12–14
Tripathi L, Singh AK, Singh S, Singh R, Chaudhary S, Sanyal I, Amla DV (2013) Optimization of regeneration and Agrobacterium-mediated transformation of immature cotyledons of chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult 113:513–527
Srivastava J, Datta S, Mishra SP (2017) Development of an efficient Agrobacterium-mediated transformation system for chickpea (Cicer arietinum L.). Biologia 72(2):153–160
Vijayan S, Kirti PB (2012) Mungbean plants expressing BjNPR1 exhibit enhanced resistance against the seedling rot pathogen, Rhizoctonia solani. Trans Res 21:193–200
Jha S, Tank HG, Prasad BD, Chattoo BB (2008) Expression of DM-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Trans Res 18:59–69
Abdallah NS, Shah D, Abbas D, Madkour M (2010) Stable integration and expression of a plant defensin in tomato confers resistance to Fusarisum wilt. GM Crops 1:344–350
Sunisha C, Sowmya HD, Usharani TR, Umesha M, Gopalkrishna HR, Saxena AK (2020) Development of stacked Antimicrobial genes in Banana for stable tolerance against Fusararium oxysporum f.sp. cubense through genetic transformation. Mol Biotech 62:8–17
Acknowledgements
The authors are grateful to the Department of Atomic Energy (DAE), Board of Research in Nuclear Sciences (BRNS), Government of India for financial assistance in the form of a project (DAE-BRNS No.2013/35/36/BRNS/1254, Dt: 30.07.2013).
Author information
Authors and Affiliations
Contributions
SKS, PJ, KKG, and VM performed all experiments. SP and VP participated in designing the experiments and writing the manuscript. SP and VP reviewed the final manuscript. VP designed and supervised the entire work. All the authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest in the present study.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sadhu, S., Jogam, P., Gande, K. et al. Expression of radish defensin (RsAFP2) gene in chickpea (Cicer arietinum L.) confers resistance to Fusarium wilt disease. Mol Biol Rep 50, 11–18 (2023). https://doi.org/10.1007/s11033-022-08021-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08021-9