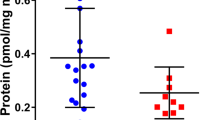
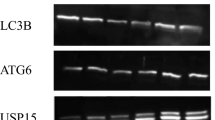
Abstract
Background
Carbonyl reductase 1 (CBR1) is a nicotinamide adenine dinucleotide phosphate (NADPH)-dependent reductase with broad substrate specificity. CBR1 catalyzes the reduction of numerous carbonyl compounds, including quinones, prostaglandins, menadione, and multiple xenobiotics, while also participating in various cellular processes, such as carcinogenesis, apoptosis, signal transduction, and drug resistance. In this study, we aimed to generate transgenic mice overexpressing mouse Cbr1 (mCbr1), characterize the mCbr1 expression in different organs, and identify changes in protein expression patterns.
Methods and Results
To facilitate a deeper understanding of the functions of CBR1, we generated transgenic mice overexpressing CBR1 throughout the body. These transgenic mice overexpress 3xFLAG-tagged mCbr1 (3xFLAG-mCbr1) under the CAG promoter. Two lines of transgenic mice were generated, one with 3xFLAG-mCbr1 expression in multiple tissues, and the other, with specific expression of 3xFLAG-mCbr1 in the heart. Pathway and network analysis using transgenic mouse hearts identified 73 proteins with levels of expression correlating with mCbr1 overexpression. The expression of voltage-gated anion channels, which may be directly related to calcium ion-related myocardial contraction, was also upregulated.
Conclusion
mCbr1 transgenic mice may be useful for further in vivo analyses of the molecular mechanisms regulated by Cbr1; such analyses will provide a better understanding of its effects on carcinogenesis and cardiotoxicity of certain cancer drugs.





Similar content being viewed by others
References
Persson B, Kallberg Y (2013) Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs). Chem Biol Interact 202:111–115. https://doi.org/10.1016/j.cbi.2012.11.009
Wermuth B (1981) Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to prostaglandin 9-ketoreductase and xenobiotic ketone reductase. J Biol Chem 256:1206–1213. https://doi.org/10.1016/S0021-9258(19)69950-3
Gonzalez-Covarrubias V, Kalabus JL, Blanco JG (2008) Inhibition of Polymorphic Human carbonyl reductase 1 (CBR1) by the cardioprotectant Flavonoid 7-monohydroxyethyl rutoside (monoHER). Pharm Res 25:1730–1734. https://doi.org/10.1007/s11095-008-9592-5
Olson LE, Bedja D, Alvey SJ, Cardounel AJ, Gabrielson KL, Reeves RH (2003) Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1. Cancer Res 63:6602–6606
Forrest GL, Gonzalez B, Tseng W, Li X, Mann J (2000) Human carbonyl reductase overexpression in the heart advances the development of doxorubicin-induced cardiotoxicity in transgenic mice. Cancer Res 60:5158–5164
Shi SM, Di L (2017) The role of carbonyl reductase 1 in drug discovery and development. Expert Opin Drug Metab Toxicol 13:859–870. https://doi.org/10.1080/17425255.2017.1356820
Hu D, Miyagi N, Arai Y, Oguri H, Miura T, Nishinaka T et al (2015) Synthesis of 8-hydroxy-2-iminochromene derivatives as selective and potent inhibitors of human carbonyl reductase 1. Org Biomol Chem 13:7487–7499. https://doi.org/10.1039/c5ob00847f
Boušová I, Skálová L, Souček P, Matoušková P (2015) The modulation of carbonyl reductase 1 by polyphenols. Drug Metab Rev 47:520–533. https://doi.org/10.3109/03602532.2015.1089885
Yokoyama Y, Xin B, Shigeto T, Umemoto M, Kasai-Sakamoto A, Futagami M et al (2007) Clofibric acid, a peroxisome proliferator-activated receptor α ligand, inhibits growth of human ovarian cancer. Mol Cancer Ther 6:1379–1386. https://doi.org/10.1158/1535-7163.MCT-06-0722
Umemoto M, Yokoyama Y, Sato S, Tsuchida S, Al-Mulla F, Saito Y (2001) Carbonyl reductase as a significant predictor of survival and lymph node metastasis in epithelial ovarian cancer. Br J Cancer 85:1032–1036. https://doi.org/10.1054/bjoc.2001.2034
Miura R, Yokoyama Y, Shigeto T, Futagami M, Mizunuma H (2015) Inhibitory effect of carbonyl reductase 1 on ovarian cancer growth via tumor necrosis factor receptor signaling. Int J Oncol 47:2173–2180. https://doi.org/10.3892/ijo.2015.3205
Kobayashi A, Yokoyama Y, Osawa Y, Miura R, Mizunuma H (2016) Gene therapy for ovarian cancer using carbonyl reductase 1 DNA with a polyamidoamine dendrimer in mouse models. Cancer Gene Ther 23:24–28. https://doi.org/10.1038/cgt.2015.61
Nishimoto Y, Murakami A, Sato S, Kajimura T, Nakashima K, Yakabe K et al (2018) Decreased carbonyl reductase 1 expression promotes tumor growth via epithelial mesenchymal transition in uterine cervical squamous cell carcinomas. Reprod Med Biol 17:173–181. https://doi.org/10.1002/rmb2.12086
Kajimura T, Sato S, Murakami A, Hayashi-Okada M, Nakashima K, Sueoka K, Sugino N (2019) Overexpression of carbonyl reductase 1 inhibits malignant behaviors and epithelial mesenchymal transition by suppressing TGF-β signaling in uterine leiomyosarcoma cells. Oncol Lett 18:1503–1512. https://doi.org/10.3892/ol.2019.10429
Takenaka K, Ogawa E, Oyanagi H, Wada H, Tanaka F (2005) Carbonyl reductase expression and its clinical significance in non-small-cell lung cancer. Cancer Epidemiol Biomarkers Prev 14:1972–1975. https://doi.org/10.1158/1055-9965.EPI-05-0060
Ikawa M, Kominami K, Yoshimura Y, Tanaka K, Nishimune Y, Okabe M (1995) Green fluorescent protein as a marker in transgenic mice. Dev Growth Differ 37:455–459. https://doi.org/10.1046/j.1440-169X.1995.t01-2-00012.x
Meng G (2018) TransgeneR: a one-stop tool for transgene integration and rearrangement discovery using sequencing data. bioRxiv. https://doi.org/10.1101/462267
Tang WH, Shilov IV, Seymour SL (2008) Nonlinear fitting method for determining local false discovery rates from decoy database searches. J Proteome Res 7:3661–3667. https://doi.org/10.1021/pr070492f
Sennels L, Bukowski-Wills JC, Rappsilber J (2009) Improved results in proteomics by use of local and peptide-class specific false discovery rates. BMC Bioinformatics 10:179. https://doi.org/10.1186/1471-2105-10-179
Xia J, Psychogios N, Young N, Wishart DS (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37:W652–W660. https://doi.org/10.1093/nar/gkp356
Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G et al (2018) MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46:W486–W494. https://doi.org/10.1093/nar/gky310
Hopp TP, Prickett KS, Price VL, Libby RT, March CJ, Pat Cerretti DP et al (1988) A short polypeptide marker sequence useful for recombinant protein identification and purification. Nat Biotechnol 6:1204–1210. https://doi.org/10.1038/nbt1088-1204
Einhauer A, Jungbauer A (2001) The FLAG™ peptide, a versatile fusion tag for the purification of recombinant proteins. J Biochem Biophys Methods 49:455–465. https://doi.org/10.1016/S0165-022X(01)00213-5
Osawa Y, Yokoyama Y, Shigeto T, Futagami M, Mizunuma H (2015) Decreased expression of carbonyl reductase 1 promotes ovarian cancer growth and proliferation. Int J Oncol 46:1252–1258. https://doi.org/10.3892/ijo.2014.2810
Oikiri H, Asano Y, Matsusaki M, Akashi M, Shimoda H, Yokoyama Y (2019) Inhibitory effect of carbonyl reductase 1 against peritoneal progression of ovarian cancer: evaluation by ex vivo 3D-human peritoneal model. Mol Biol Rep 46:4685–4697. https://doi.org/10.1007/s11033-019-04788-6
Miyoshi I, Takahashi K, Kon Y, Okamura T, Mototani Y, Araki Y, Kasai N (2002) Mouse transgenic for murine oviduct-specific glycoprotein promoter-driven simian virus 40 large T-antigen: tumor formation and its hormonal regulation. Mol Reprod Dev 63:168–176. https://doi.org/10.1002/mrd.10175
Piska K, Koczurkiewicz P, Bucki A, Wójcik-Pszczoła K, Kołaczkowski M, Pękala E (2017) Metabolic carbonyl reduction of anthracyclines - role in cardiotoxicity and cancer resistance. Reducing enzymes as putative targets for novel cardioprotective and chemosensitizing agents. Investig New Drugs 35:375–385. https://doi.org/10.1007/s10637-017-0443-2
Zhong L, Shen H, Huang C, Jing H, Cao D (2011) AKR1B10 induces cell resistance to daunorubicin and idarubicin by reducing C13 ketonic group. Toxicol Appl Pharmacol 255:40–47. https://doi.org/10.1016/j.taap.2011.05.014
Matsunaga T, Wada Y, Endo S, Soda M, El-Kabbani O, Hara A (2012) Aldo-keto reductase 1B10 and its role in proliferation capacity of drug-resistant cancers. Front Pharmacol 3:5. https://doi.org/10.3389/fphar.2012.00005
Stolarz AJ, Sarimollaoglu M, Marecki JC, Fletcher TW, Galanzha EI, Rhee SW et al (2019) Doxorubicin activates ryanodine receptors in rat lymphatic muscle cells to attenuate rhythmic contractions and lymph flow. J Pharmacol Exp Ther 371:278–289. https://doi.org/10.1124/jpet.119.257592
Arai Y, Endo S, Miyagi N, Abe N, Miura T, Nishinaka T et al (2015) Structure–activity relationship of flavonoids as potent inhibitors of carbonyl reductase 1 (CBR1). Fitoterapia 101:51–56. https://doi.org/10.1016/j.fitote.2014.12.010
Carlquist M, Frejd T, Gorwa-Grauslund MF (2008) Flavonoids as inhibitors of human carbonyl reductase 1. Chem Biol Interact 174:98–108. https://doi.org/10.1016/j.cbi.2008.05.021
Zemanova L, Hofman J, Novotna E, Musilek K, Lundova T, Havrankova J et al (2015) Flavones inhibit the activity of AKR1B10, a promising therapeutic target for cancer treatment. J Nat Prod 78:2666–2674. https://doi.org/10.1021/acs.jnatprod.5b00616
Huang W, Ding L, Huang Q, Hu H, Liu S, Yang X et al (2010) Carbonyl reductase 1 as a novel target of (-)-epigallocatechin gallate against hepatocellular carcinoma. Hepatology 52:703–714. https://doi.org/10.1002/hep.23723
Ito Y, Mitani T, Harada N, Isayama A, Tanimori S, Takenaka S et al (2013) Identification of carbonyl reductase 1 as a resveratrol-binding protein by affinity chromatography using 4′-amino-3,5-dihydroxy-trans-stilbene. J Nutr Sci Vitaminol (Tokyo) 59:358–364. https://doi.org/10.3177/jnsv.59.358
Hintzpeter J, Hornung J, Ebert B, Martin HJ, Maser E (2015) Curcumin is a tight-binding inhibitor of the most efficient human daunorubicin reductase - carbonyl reductase 1. Chem Biol Interact 234:162–168. https://doi.org/10.1016/j.cbi.2014.12.019
Hintzpeter J, Seliger JM, Hofman J, Martin HJ, Wsol V, Maser E (2016) Inhibition of human anthracycline reductases by emodin - A possible remedy for anthracycline resistance. Toxicol Appl Pharmacol 293:21–29. https://doi.org/10.1016/j.taap.2016.01.003
Gambliel HA, Burke BE, Cusack BJ, Walsh GM, Zhang YL, Mushlin PS, Olson RD (2002) Doxorubicin and C-13 deoxydoxorubicin effects on ryanodine receptor gene expression. Biochem Biophys Res Commun 291:433–438. https://doi.org/10.1006/bbrc.2002.6380
Hiller S, Abramson J, Mannella C, Wagner G, Zeth K (2010) The 3D structures of VDAC represent a native conformation. Trends Biochem Sci 35:514–521. https://doi.org/10.1016/j.tibs.2010.03.005
Freeland MM, Angulo J, Davis AL, Flook AM, Garcia BL, King NA et al (2012) Sex differences in improved efficacy of doxorubicin chemotherapy in Cbr1+/– mice. Anticancer Drugs 23:584–589. https://doi.org/10.1097/CAD.0b013e3283512726
Arima-Yoshida F, Raveau M, Shimohata A, Amano K, Fukushima A, Watanave M et al (2020) Impairment of spatial memory accuracy improved by Cbr1 copy number resumption and GABAB receptor-dependent enhancement of synaptic inhibition in Down syndrome model mice. Sci Rep 10:14187. https://doi.org/10.1038/s41598-020-71085-9
Deutsch EW, Csordas A, Sun Z, Jarnuczak A, Perez-Riverol Y, Ternent T et al (2017) The ProteomeXchange Consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res 45:D1100–D1106. https://doi.org/10.1093/nar/gkw936
Okuda S, Watanabe Y, Moriya Y, Kawano S, Yamamoto T, Matsumoto M et al (2017) JPOSTrepo: an international standard data repository for proteomes. Nucleic Acids Res 45:D1107–D1111. https://doi.org/10.1093/nar/gkw1080
Acknowledgements
The authors thank Hiroshi Kijima for his assistance with the immunohistochemistry analysis; Takeshi Shimizu, Shoko Nagata, and Hirotaka Fujita for their technical assistance; and Miyu Miyazaki for her technical support in the mass spectrometry analysis.
Author information
Authors and Affiliations
Contributions
M.Y., H.F., and Y.Y. conceived the project; T.F. and H.F. designed and supervised the project; Y.T. performed the LC-MS/MS analysis; D.M. performed the NGS analysis; M.I. generated the transgenic mice; M.Y. and Y.K. performed the other experiments; and M.Y., T.F., H.F., and Y.Y. wrote the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Funding
None.
Competing Interests:
Y.Y. and M.Y. have filed a patent application for the transgenic mice with details as follows: Name: Transgenic non-human animals; Number: Japanese Patent Application No. 2022-009381; Status: Under review; Specific aspect of manuscript covered in the patent application: All the information reported in this study.
Ethics approval:
All animal experiments were approved by the Institutional Animal Care and Use Committee at the Research Institute for Microbial Diseases, Osaka University (4111) and Hirosaki University (M19024). This study complied with the ARRIVE guidelines. All methods were performed in accordance with the relevant guidelines and regulations.
Consent to participate:
Not applicable.
Consent to publish:
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yokoyama, M., Fujita, T., Kadonosawa, Y. et al. Development of transgenic mice overexpressing mouse carbonyl reductase 1. Mol Biol Rep 50, 531–540 (2023). https://doi.org/10.1007/s11033-022-07994-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07994-x