Abstract
Background
Molecular studies on egg production in ducks were mostly focused on brain and ovaries as they are directly involved in egg production. Liver plays a vital role in cellular lipid metabolism. It also plays a decisive role in reproductive organ development, including yolk generation in laying ducks at sexual maturity. However, the precise molecular mechanism involved in the liver-blood-ovary axis in ducks remains elusive.
Methods and Results
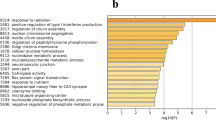
In this study, we analysed the liver transcriptome of laying (LA), immature (IM) and broody (BR) ducks using RNA sequencing to understand the role of genes expressed in the liver. The comparative transcriptome analysis revealed 82 DEGs between LA and IM ducks, 47 DEGs between LA and BR ducks and 51 DEGs between IM and BR ducks. GO analysis of DEGs, showed that DEGs were mainly involved in cellular anatomical entity, intracellular, metabolic process, and binding. Furthermore, pathway analysis indicated the important role of Wnt signaling pathway in egg formation and embryo development. Our study showed several candidate genes including vitellogenin-1, vitellogenin-2, riboflavin binding protein, G protein subunit gamma 4, and fatty acid binding protein 3 that are potentially related to egg production in ducks.
Conclusions
The study provides valuable information on the genes responsible for egg production and thus, pave the way for further investigation on the molecular mechanisms of egg production in duck.



Similar content being viewed by others
Data availability statement
The transcriptome data are available at NCBI under the accession number PRJNA791524.
References
Padhi MK (2016) Importance of Indigenous Breeds of Chicken for Rural Economy and Their Improvements for Higher Production Performance. Scientifica 2016:1–9. https://doi.org/10.1155/2016/2604685
Kumar M, Dahiya SP, Ratwan P (2021) Backyard poultry farming in India: A tool for nutritional security and women empowerment. Biol Rhythm Res 52:1476–1491. https://doi.org/10.1080/09291016.2019.1628396
Zhu Z, Miao Z, Chen H, Xin Q, Li L, Lin R et al (2016) Ovarian transcriptomic analysis of Shan Ma ducks at peak and late stages of egg production. Asian-Australas J Anim Sci 30:1215–1224. https://doi.org/10.5713/ajas.16.0470
George TG, Nayar R, Cyriac S (2012) Yields and Ratios of Different Meat Parts of Vigova Super M and Kuttanad Ducks: A Comparison. Int J Sci Res 3:2817–2819
Ren J, Sun C, Chen L, Hu J, Huang X, Liu X et al (2019) Exploring differentially expressed key genes related to development of follicle by RNA-seq in Peking ducks (Anas platyrhynchos). PLoS ONE 14:e0209061. https://doi.org/10.1371/journal.pone.0209061
Mishra SK, Chen B, Zhu Q, Xu Z, Ning C, Yin H et al (2020) Transcriptome analysis reveals differentially expressed genes associated with high rates of egg production in chicken hypothalamic-pituitary-ovarian axis. Sci Rep 10:5976. https://doi.org/10.1038/s41598-020-62886-z
Ma Z, Jiang K, Wang D, Wang Z, Gu Z, Li G et al (2021) Comparative analysis of hypothalamus transcriptome between laying hens with different egg-laying rates. Poult Sci 101110. https://doi.org/10.1016/j.psj.2021.101110
Zhang T, Chen L, Han K, Zhang X, Zhang G, Dai G et al (2019) Transcriptome analysis of ovary in relatively greater and lesser egg producing Jinghai Yellow Chicken. Ani Rep Sci 208:106114. https://doi.org/10.1016/j.anireprosci.2019.106114
Yu J, Lou Y, Zhao A (2016) Transcriptome analysis of follicles reveals the importance of autophagy and hormones in regulating broodiness of Zhedong white goose. Sci Rep 6:1–12. https://doi.org/10.1038/srep36877
Li H, Wang T, Xu C, Wang D, Ren J, Li Y et al (2015) Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genomics 16:763. http://www.biomedcentral.com/1471-2164/16/763
Alvarenga R, Zangeronimo M, Pereira L, Rodrigues P, Gomide E (2011) Lipoprotein metabolism in poultry. Worlds Poult Sci J 67:431–440. https://doi.org/10.1017/S0043933911000481
Li H, Gu Z, Yang L, Tian Y, Kang X, Liu X (2018) Transcriptome profile analysis reveals an estrogen induced LncRNA associated with lipid metabolism and carcass traits in chickens (Gallus Gallus). Cell Physiol Biochem 50:1638–1658. https://doi.org/10.1159/00049478
Zheng A, Liu G, Zhang Y, Hou S, Chang W, Zhang S et al (2012) Proteomic analysis of liver development of lean Pekin duck (Anas platyrhynchos domestica). J Proteom 75:5396–5413. https://doi.org/10.1016/j.jprot.2012.06.019
Andrews S (2010) FastQC: A quality control tool for high throughput sequence data. http://www.bioinformaticsbabrahamacuk/projects/fastqc/
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. https://doi.org/10.14806/ej.17.1.200
Bray NL, Pimentel H, Melsted P, Pachter L (2016) Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. https://www.nature.com/articles/nbt.3519
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mi H, Ebert D, Muruganujan A, Mills C, Albou L-P, Mushayamaha T et al (2021) PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucl Aci Res 49(D1):D394–403. https://doi.org/10.1093/nar/gkaa1106
Gloux A, Duclos MJ, Brionne A, Bourin M, Nys Y, Réhault-Godbert S (2019) Integrative analysis of transcriptomic data related to the liver of laying hens: from physiological basics to newly identified functions. BMC Genomics 20:821. https://doi.org/10.1186/s12864-019-6185-0
Johnson AL (2015) Chap. 28 - Reproduction in the Female. In: Scanes CG, editor. Sturkie’s Avian Physiology (Sixth Edition) San Diego: Academic Press, pp 635 – 65. https://www.sciencedirect.com/science/article/pii/B9780124071605000282
Nimpf J, Schneider WJ (1991) Receptor-Mediated Lipoprotein Transport in Laying Hens. J Nutr 121:1471–1474. https://doi.org/10.1093/jn/121.9.1471
Schneider WJ (1995) Yolk precursor transport in the laying hen. Currs Opin Lipidol 6:92–96. https://doi.org/10.1097/00041433-199504000-00006
Montorzi M, Falchuk KH, Vallee BL (1995) Vitellogenin and lipovitellin: zinc proteins of Xenopus laevis oocytes. Biochemistry 34:10851–10858. https://doi.org/10.1021/bi00034a018
Yen CF, Lin EC, Wang YH, Wang PH, Lin HW, Hsu JC et al (2009) Abundantly expressed hepatic genes and their differential expression in liver of prelaying and laying geese. Poult Sci 88:1955–1962. https://doi.org/10.3382/ps.2008-00473
Ding ST, Yen CF, Wang PH, Lin HW, Hsu JC, Shen TF (2007) The Differential Expression of Hepatic Genes Between Prelaying and Laying Geese. Poult Sci 86:1206–1212. https://doi.org/10.1093/ps/86.6.1206
Zabłocka A, Bobak Ł, Macała J, Rymaszewska J, Kazana W, Zambrowicz A (2021) Comparative Studies of Yolkin Preparations Isolated from Egg Yolks of Selected Bird Species. Chem Biodivers 18:e2100178. https://doi.org/10.1002/cbdv.202100178
Tian X, Gautron J, Monget P, Pascal G (2010) What makes an egg unique? Clues from evolutionary scenarios of egg-specific genes. Biol Reprod 83:893–900. https://doi.org/10.1095/biolreprod.110.085019
Bourin M, Gautron J, Berges M, Hennequet-Antier C, Cabau C, Nys Y et al (2012) Transcriptomic profiling of proteases and antiproteases in the liver of sexually mature hens in relation to vitellogenesis. BMC Genomics 13:457. https://doi.org/10.1186/1471-2164-13-457
Rasikh AH (2019) Role of vitamins in animal health and production. Int Int J Vet Sci Anim Husb 4:40–43
White IIIHB (1996) Sudden death of chicken embryos with hereditary riboflavin deficiency. J Nutr 126. https://doi.org/10.1093/jn/126.suppl_4.1303S. 1303S-1307S
Tang J, Hu J, Xue M et al (2019) Maternal diet deficient in riboflavin induces embryonic death associated with alterations in the hepatic proteome of duck embryos. Nutr Metab 16:19. https://doi.org/10.1186/s12986-019-0345-8
Atshaves BP, McIntosh AM, Lyuksyutova OI, Zipfel W, Webb WW, Schroeder F (2004) Liver fatty acid-binding protein gene ablation inhibits branched-chain fatty acid metabolism in cultured primary hepatocytes. J Biol Chem 279:30954–30965. https://doi.org/10.1074/jbc.M313571200
Martin GG, Danneberg H, Kumar LS, Atshaves BP, Erol E, Bader M et al (2003) Decreased liver fatty acid binding capacity and altered liver lipid distribution in mice lacking the liver fatty acid-binding protein gene. J Biol Chem 278:21429–21438. https://doi.org/10.1074/jbc.M300287200
Kaikaus RM, Sui Z, Lysenko N, Wu NY, de Montellano PO, Ockner RK et al (1993) Regulation of pathways of extramitochondrial fatty acid oxidation and liver fatty acid-binding protein by long-chain monocarboxylic fatty acids in hepatocytes. Effect of inhibition of carnitine palmitoyltransferase I. J Biol Chem 268:26866–26871. https://doi.org/10.1016/S0021-9258(19)74191-X
Woodford JK, Behnke WD, Schroeder F (1995) Liver fatty acid binding protein enhances sterol transfer by membrane interaction. Mol Cell Biochem 152:51–62. https://doi.org/10.1007/BF01076463
Fan R, Cao Z, Chen M, Wang H, Liu M, Gao M et al (2021) Effects of the FABP4 gene on steroid hormone secretion in goose ovarian granulosa cells. Bri Poult Sci 62:81–91. https://doi.org/10.1080/00071668.2020.1817325
Cao Z, Meng B, Fan R, Liu M, Gao M, Xing Z et al (2018) Comparative proteomic analysis of ovaries from Huoyan geese between pre-laying and laying periods using an iTRAQ-based approach. Poult Sci 97:2170–2182. https://doi.org/10.3382/ps/pey029
Gong Y-N, Li W-W, Sun J-L, Ren F, He L, Jiang H et al (2010) Molecular cloning and tissue expression of the fatty acid-binding protein (Es-FABP) gene in female Chinese mitten crab (Eriocheir sinensis). BMC Mol Bio 11:71. https://doi.org/10.1186/1471-2199-11-71
Iseki S, Amano O, Fujii H, Kanda T, Ono T (1995) Immunohiostochemical localization of two types of fatty acid-binding proteins in rat ovaries during postnatal development and in immature rat ovaries treated with gonadotropins. Anat Rec 241:235–243. https://doi.org/10.1002/ar.1092410210
Ko YH, Cheng CH, Shen TF, Ding ST (2004) Cloning and expression of Tsaiya duck liver fatty acid binding protein. Poult Sci 83:1832–1838. https://doi.org/10.1093/ps/83.11.1832
Murai A, Furuse M, Kitaguchi K, Kusumoto K, Nakanishi Y, Kobayashi M et al (2009) Characterization of critical factors influencing gene expression of two types of fatty acid-binding proteins (L-FABP and Lb-FABP) in the liver of birds. Comp Biochem Physiol Part A Mol Integr Physiol 154:216–223. https://doi.org/10.1016/j.cbpa.2009.06.007
Yang S, Suh Y, Choi YM, Shin S, Han JY, Lee K (2013) Loss of fat with increased adipose triglyceride lipase-mediated lipolysis in adipose tissue during laying stages in quail. Lipids 48:13–21. https://doi.org/10.1007/s11745-012-3742-6
Nie R, Zheng X, Zhang W, Zhang B, Ling Y, Zhang H, Wu C (2022) Morphological Characteristics and Transcriptome Landscapes of Chicken Follicles during Selective Development. Animals 12: 713. https://doi.org/10.3390/ani12060713
Ren J, Du X, Zeng T, Chen L, Shen J, Lu L et al (2017) Divergently expressed gene identification and interaction prediction of long noncoding RNA and mRNA involved in duck reproduction. Ani Rep Sci 185:8–17. https://doi.org/10.1016/j.anireprosci.2017.07.012
Pertiñez SP, Wilson PW, Icken W, Cavero D, Bain MM, Jones AC et al (2020) Transcriptome analysis of the uterus of hens laying eggs differing in cuticle deposition. BMC Genomics 21:1–15. https://doi.org/10.1186/s12864-020-06882-7
Khan S, Wu S-B, Roberts J (2019) RNA-sequencing analysis of shell gland shows differences in gene expression profile at two time-points of eggshell formation in laying chickens. BMC Genomics 20:1–20. https://doi.org/10.1186/s12864-019-5460-4
Wan Y, Jin S, Ma C, Wang Z, Fang Q, Jiang R (2017) RNA-Seq reveals seven promising candidate genes affecting the proportion of thick egg albumen in layer-type chickens. Sci Rep 7:1–9. https://doi.org/10.1038/s41598-017-18389-5
Acknowledgements
The first author is thankful to the Central University of Kerala for the support in the form of a research fellowship. The corresponding author is thankful to the Central University of Kerala for providing the necessary facilities to carry out the work.
Funding
The study was partially supported by the Department of Biotechnology (BT/PR14584/AAQ/1/647/2015), New Delhi and Science and Engineering Research Board (EEQ/2016/000390), Department of Science and Technology, New Delhi.
Author information
Authors and Affiliations
Contributions
MN: conceived the study. KN: performed the experiments. KN, KS, DV, DN: performed the analyses. KN, M.N, KS, SS. wrote the manuscript. All the authors have seen and approved the manuscript being submitted.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical approval
There were no experiments on duck in the present study. The liver tissue samples were collected from the deceased domestic ducks from a slaughterhouse in compliance with the research ethical standards of India. Hence no approval of animal ethics committee was required for this study.
Consent to publish
All authors have significantly contributed to the study, read the manuscript, and consented to submit for publication in the journal Molecular Biology Reports.
Consent to participate
This study does not contain any studies with human participants.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nimisha, K., Srikanth, K., Velayutham, D. et al. Comparative liver transcriptome analysis of duck reveals potential genes associated with egg production. Mol Biol Rep 49, 5963–5972 (2022). https://doi.org/10.1007/s11033-022-07380-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07380-7