Abstract
Background
Extracellular vesicles (EVs) contain thousands of proteins and nucleic acids, playing an important role in cell–cell communications. Sertoli cells have been essential in the testis as a “nurse cell”. However, EVs derived from human Sertoli cells (HSerCs) have not been well investigated.
Methods
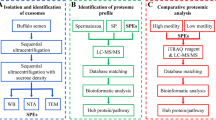
EVs were isolated from HSerCs via ultracentrifugation and characterized by transmission electron microscopy, tunable resistive pulse sensing, and Western blotting. The cargo carried by HSerCs-EVs was measured via liquid chromatography-mass spectrometry and GeneChip miRNA Arrays. Bioinformatic analysis was performed to reveal potential functions of HSerCs-EVs.
Results
A total of 860 proteins with no less than 2 unique peptides and 88 microRNAs with high signal values were identified in HSerCs-EVs. Biological processes related to molecular binding, enzyme activity, and regulation of cell cycle were significantly enriched. Specifically, many proteins in HSerCs-EVs were associated with spermatogenesis and regulation of immune system, including Septins, Large proline-rich protein BAG6, Clusterin, and Galectin-1. Moreover, abundant microRNAs within HSerCs-EVs (miR-638, miR-149-3p, miR-1246, etc.) had a possible impact on male reproductive disorders such as asthenozoospermia and oligozoospermia.
Conclusions
Our study has shown that HSerCs-EVs contain diverse components such as proteins and microRNAs. Further research is required to evaluate HSerCs-EVs in spermatogenesis, which are underutilized but highly potent resources with particular promise for male infertility.


Similar content being viewed by others
Abbreviations
- EVs:
-
Extracellular vesicles
- SCs:
-
Sertoli cells
- HSerCs:
-
Human Sertoli cells
- LC:
-
Liquid chromatography
- miRNA:
-
MicroRNA
- MS:
-
Mass spectrometry
- PBS:
-
Phosphate-buffered saline
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
References
Kalluri R, LeBleu VS (2020) The biology, function, and biomedical applications of exosomes. Science 6478:6977
Yáñez-Mó M, Siljander PRM, Andreu Z et al (2015) Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 1:27066
França LR, Hess RA, Dufour JM, Hofmann MC, Griswold MD (2016) The Sertoli cell: one hundred fifty years of beauty and plasticity. Andrology 2:189–212
Kaur G, Thompson LA, Dufour JM (2014) Sertoli cells: immunological sentinels of spermatogenesis. Semin Cell Dev Biol 1:36–44
Zhao L, Yao C, Xing X et al (2020) Single-cell analysis of developing and azoospermia human testicles reveals central role of Sertoli cells. Nat Commun 1:5683
Meroni SB, Galardo MN, Rindone G, Gorga A, Riera MF, Cigorraga SB (2019) Molecular mechanisms and signaling pathways involved in Sertoli cell proliferation. Front Endocrinol 10:224
Guo J, Sosa E, Chitiashvili T et al (2021) Single-cell analysis of the developing human testis reveals somatic niche cell specification and fetal germline stem cell establishment. Cell Stem Cell 4:764–778
Urriola-Muñoz P, Lizama C, Lagos-Cabré R, Reyes JG, Moreno RD (2014) Differential expression and localization of ADAM10 and ADAM17 during rat spermatogenesis suggest a role in germ cell differentiation. Biol Res 1:31
Huang LS, Voyiaziakis E, Chen HL, Rubin EM, Gordon JW (1996) A Novel functional role for apolipoprotein B in male infertility in heterozygous apolipoprotein B knockout mice. Proc Natl Acad Sci USA 20:10903–10907
Okunade GW, Miller ML, Pyne GJ et al (2004) Targeted ablation of plasma membrane Ca2+-ATPase (PMCA) 1 and 4 indicates a major housekeeping function for PMCA1 and a critical role in hyperactivated sperm motility and male fertility for PMCA4. J Biol Chem 32:33742–33750
Bromfield E, Aitken RJ, Nixon B (2015) Novel characterization of the HSPA2-stabilizing protein BAG6 in human spermatozoa. Mol Hum Reprod 10:755–769
Intasqui P, Agarwal A, Sharma R, Samanta L, Bertolla RP (2018) Towards the identification of reliable sperm biomarkers for male infertility: a sperm proteomic approach. Andrologia 3:1–10
Nakamura M, Moriya M, Baba T et al (1993) An endoplasmic reticulum protein, calreticulin, is transported into the acrosome of rat sperm. Exp Cell Res 1:101–110
Jenkins ZA, Hååg PG, Johansson HE (2001) Human eIF5A2 on chromosome 3Q25-Q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5a with tissue-specific expression. Genomics 1:101–109
Testa E, Nardozi D, Antinozzi C et al (2018) H2AFX and MDC1 promote maintenance of genomic integrity in male germ cells. J Cell Sci 6:1–10
Yuen BT, Bush KM, Barrilleaux BL, Cotterman R, Knoepfler PS (2014) Histone H33 regulates dynamic chromatin states during spermatogenesis. Development 18:3483–3494
Yao GD, Shi SL, Song WY et al (2015) Role of PAFAH1B1 in human spermatogenesis, fertilization and early embryonic development. Reprod Biomed Online 5:613–624
Shi H, Liu J, Zhu P et al (2018) Expression of peroxiredoxins in the human testis, epididymis and spermatozoa and their role in preventing H2O2-induced damage to spermatozoa. Folia Histochem Cytobiol 3:141–150
Nguyen TM (2017) Impact of 5’-amp-activated protein kinase on male gonad and spermatozoa functions. Front Cell Dev Biol 25:1–10
Lin YH, Kuo YC, Chiang HS, Kuo PL (2011) The role of the septin family in spermiogenesis. Spermatogenesis 4:298–302
Muñoz X, Mata A, Bassas L, Larriba S (2015) Altered miRNA signature of developing germ-cells in infertile patients relates to the severity of spermatogenic failure and persists in spermatozoa. Sci Rep 5:17991
Salas-Huetos A, Blanco J, Vidal F et al (2016) Spermatozoa from normozoospermic fertile and infertile individuals convey a distinct miRNA Cargo. Andrology 6:1028–1036
Belleannée C, Légaré C, Calvo E, Thimon V, Sullivan R (2013) MicroRNA signature is altered in both human epididymis and seminal microvesicles following vasectomy. Hum Reprod 6:1455–1467
Zhou R, Zhang Y, Du G et al (2018) Down-regulated let-7b-5p represses glycolysis metabolism by targeting AURKB in asthenozoospermia. Gene 663:83–87
Salas-Huetos A, Blanco J, Vidal F et al (2015) Spermatozoa from patients with seminal alterations exhibit a differential micro-ribonucleic acid profile. Fertil Steril 3:591–601
Lian J, Zhang X, Tian H et al (2009) Altered microRNA expression in patients with non-obstructive azoospermia. Reprod Biol Endocrinol 13:7
Wu W, Qin Y, Li Z et al (2013) Genome-wide microRNA expression profiling in idiopathic non-obstructive azoospermia: significant up-regulation of mir-141, miR-429 and miR-7-1-3p. Hum Reprod 7:1827–1836
Zhang J, Liu Q, Zhang W et al (2010) Comparative profiling of genes and miRNAs expressed in the newborn, young adult, and aged human epididymides. Acta Biochim Biophys Sin 2:145–153
Yao C, Yuan Q, Niu M et al (2017) Distinct expression profiles and novel targets of microRNAs in human spermatogonia, pachytene spermatocytes, and round spermatids between OA patients and NOA patients. Mol Ther Nucleic Acids 9:182–194
Hu L, Wu C, Guo C, Li H, Xiong C (2014) Identification of microRNAs predominately derived from testis and epididymis in human seminal plasma. Clin Biochem 10–11:967–972
Tang D, Huang Z, He X et al (2018) Altered miRNA profile in testis of post-cryptorchidopexy patients with non-obstructive azoospermia. Reprod Biol Endocrinol 1:78
Mruk DD, Cheng CY (2015) The mammalian blood-testis barrier: its biology and regulation. Endocr Rev 5:564–591
Mancuso F, Calvitti M, Milardi D et al (2018) Testosterone and FSH modulate Sertoli cell extracellular secretion: proteomic analysis. Mol Cell Endocrinol 476:1–7
Salas-Huetos A, James ER, Aston KI, Carrell DT, Jenkins TG, Yeste M (2020) The role of miRNAs in male human reproduction: a systematic review. Andrology 1:7–26
Vojtech L, Woo S, Hughes S et al (2014) Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 11:7290–7304
Liu J, Schiltz JF, Ashar HR, Chada KK (2003) Hmga1 is required for normal sperm development. Mol Reprod Dev 1:81–89
Xu HY, Zhang HX, Xiao Z, Qiao J, Li R (2019) Regulation of anti-müllerian hormone (AMH) in males and the associations of serum AMH with the disorders of male fertility. Asian J Androl 2:109–114
Bentson LF, Agbor VA, Agbor LN et al (2013) New point mutation in golga3 causes multiple defects in spermatogenesis. Andrology 3:440–450
Acknowledgements
The authors would like to thank the donor of human Sertoli cells, who made this study possible. We also appreciate the Human Protein Atlas and the PRIDE, GEO, GO, KEGG databases for providing their platforms as well as contributors for uploading their meaningful datasets.
Funding
The work was supported by the Horizontal Projects and the National Natural Science Foundation of China (Grant No. 81401194).
Author information
Authors and Affiliations
Contributions
Conception and design: R-LG, Y-MY. Administrative support: X-SL, Z-CX, SWK, WJB. Collection and assembly of data: X-HT, S-JG, W-JT, W-PS, Y-YG. Data analysis and interpretation: X-HT, S-JG. Manuscript writing: X-HT, S-JG. Final approval of manuscript: All authors.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
Ethical approval was waived since this article does not contain any studies with human participants or animals. Human Sertoli cells were commercially available and purchased from ScienCell Research Laboratories (Cat. No. 4520; Carlsbad, CA, USA).
Data availability
All data generated or analyzed during this study are included in this published article and its supplementary information files, which are available on reasonable request. The microarray data have been available in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE192696. Raw mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030640.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tan, XH., Gu, SJ., Tian, WJ. et al. Extracellular vesicles derived from human Sertoli cells: characterizations, proteomic analysis, and miRNA profiling. Mol Biol Rep 49, 4673–4681 (2022). https://doi.org/10.1007/s11033-022-07316-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-07316-1