Abstract
Background
Soybean is largely grown and considered among the top oilseed crops. Three Pakistani cultivars, NARC-II (N), Swat-84 (S), and Rawal-I (R) were employed for RNA-Seq based transcriptome analysis to explore their genetic potential and performance in our local environment.
Methods and results
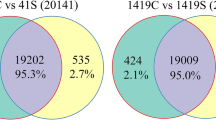
We grew the plants in glass house at same conditions and sampled leaves for RNA-Seq analysis in triplicate for each variety. We retrieved 2225 differentially expressed genes (DEGs) between S vs R, 2591 DEGs between S vs N, and 1221 DEGs between R vs N cultvars. These genes consist of transcription factors representing Basic Helix-loop Helix, myeloblastosis, ethylene response factors, and WRKY amino acid motif (WRKY) type major families that were up-regulated. KEGG pathway analysis revealed that MAPK, plant hormone signal transduction, and Phenylpropanoid biosynthesis pathways were the most dominant pathways involved in plant defense and growth. Comparative analysis showed that Swat-84 (S) cultivar had better gene expression among these varieties having higher number of DEGs, where mostly genes related to important phenotypic traits were up regulated.
Conclusions
This is a pilot study to investigate and functionally characterise the DEG involved in the stress response in the cultivars studied.




Similar content being viewed by others
Data availability
All RNA-Seq reads have been submitted to the NCBI Sequence Read Archive (SRA) database under accession number PRJNA718712.
References
SoyStats. American Soybean Association (ASA) (2019) http://soystats.com/international-world-oilseed-production/. Accessed 26 Mar 2021
Khurshid H et al (2017) Miracle crop: the present and future of soybean production in Pakistan. MOJ Biol Med 2(1):189–191
Selle PH et al (2020) Synthetic and crystalline amino acids: alternatives to soybean meal in chicken-meat production. Animals (Basel) 10(4):729
Ullah A et al (2021) Assessment of phenotypic and molecular diversity in soybean [Glycine max (L.) Merr.] germplasm using morpho-biochemical attributes and SSR markers. Genet Resourc Crop Evolut 68:2827
Anwar Malik MF et al (2016) Analysis of genetic diversity of soybean germplasm from five different origins using RAPD markers. Acta Agric Scand Sect B Soil Plant Sci 67(2):148–154
Pakistan: Oilseeds and Products Annual (2020) https://www.fas.usda.gov/data/pakistan-oilseeds-and-products-annual-5. Accessed 27 Mar 2021
Asad SA (2020) Agriculture: soya bean a miracle crop for national food security. https://www.dawn.com/news/1527911. Accessed 27 Mar 2021
Dong Y et al (2001) The genetic diversity of annual wild soybeans grown in China. Theor Appl Genet 103(1):98–103
Anwar Malik MF et al (2017) Analysis of genetic diversity of soybean germplasm from five different origins using RAPD markers. Acta Agric Scand Sect B 67(2):148–154
Iqbal Z et al (2010) Genetic divergence and correlation studies of soybean [Glycine max (L.) Merrill.] genotypes. Genet Mol Biol 42(2):971–976
Vidal RO et al (2012) Identification of SNPs in RNA-seq data of two cultivars of Glycine max (soybean) differing in drought resistance. Genet Mol Biol 35(1 suppl):331–334
Du H et al (2019) RNA-Seq analysis reveals transcript diversity and active genes after common cutworm (Spodoptera litura Fabricius) attack in resistant and susceptible wild soybean lines. BMC Genomics 20(1):237
Baig DD (2021) Soybean. http://www.parc.gov.pk/index.php/en/csi/137-narc/crop-sciences-institue/731-soybean. Accessed 19 May 2021
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17(1):10–12
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930
Mortazavi A et al (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5(7):621–628
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Kolberg L et al (2020) gprofiler2—an R package for gene list functional enrichment analysis and namespace conversion toolset g: Profiler. F1000Research 9:709
Reimand J et al (2016) g: Profiler—a web server for functional interpretation of gene lists (2016 update). Nucleic Acid Res 44(W1):W83–W89
Yu GJB (2018) clusterProfiler: universal enrichment tool for functional and comparative study, p. 256784
Du J et al (2014) KEGG-PATH: Kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol BioSyst 10(9):2441–2447
Zheng Y et al (2016) iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant 9(12):1667–1670
Gharibi S et al (2019) The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech.f. Phytochemistry 162:90–98
Schroeter H et al (2002) MAPK signaling in neurodegeneration: influences of flavonoids and of nitric oxide. Neurobiol Aging 23(5):861–880
Selmar D (2008) Potential of salt and drought stress to increase pharmaceutical significant secondary compounds in plants. Landbauforschung Volkenrode 58(1/2):139
Ma D et al (2014) Expression of flavonoid biosynthesis genes and accumulation of flavonoid in wheat leaves in response to drought stress. Plant Physiol Biochem 80:60–66
Sharma A et al (2016) Pre-sowing seed treatment with 24-epibrassinolide ameliorates pesticide stress in Brassica juncea L. through the modulation of stress markers. Front Plant Sci 7:1569
Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 5:170
Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4:248
Moffat CS et al (2012) ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7(4):e35995
Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379(6):633–646
Huang Z et al (2017) APETALA 2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol 215(3):1197–1209
Ohto MA et al (2009) Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod 22(4):277–289
Johnson ET, Dowd PF (2004) Differentially enhanced insect resistance, at a cost, in Arabidopsis thaliana constitutively expressing a transcription factor of defensive metabolites. J Agric Food Chem 52(16):5135–5138
Mengiste T et al (2003) The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15(11):2551–2565
Soler M et al (2015) The Eucalyptus grandis R2R3-MYB transcription factor family: evidence for woody growth-related evolution and function. New Phytol 206(4):1364–1377
Koini MA et al (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19(5):408–413
Franklin KA et al (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108(50):20231–20235
Guan Y et al (2014) Phosphorylation of a WRKY transcription factor by MAPKs is required for pollen development and function in Arabidopsis. PLoS Genet 10(5):e1004384
Dilkes BP et al (2008) The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol 6(12):2707–2720
Grunewald W et al (2013) Tightly controlled WRKY23 expression mediates Arabidopsis embryo development. EMBO Rep 14(12):1136–1142
Rinerson CI et al (2015) The WRKY transcription factor family and senescence in switchgrass. BMC Genomics 16(1):912
Zheng Z et al (2006) Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Planr J 48(4):592–605
Mao G et al (2011) Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23(4):1639–1653
Qi T et al (2014) Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26(3):1118–1133
Funding
Higher Education Commission (HEC), Pakistan has provided funds through its NRPU Project No. 7838/Balochistan/NRPU/R&D/HEC/2017.
Author information
Authors and Affiliations
Contributions
ZJ and WH designed and executed the experiment. KN grew the plants and did sampling for RNA-Seq, AT and WA did data analysis. HA and SF contributed to manuscript preparation and review.
Corresponding author
Ethics declarations
Conflict of interest
The authors decolor no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2021_7104_MOESM2_ESM.pptx
Supplementary Fig. S1 An overview of regulated TFs in three comparisons. a Differentially regulated TFs show that mostly TFs are upregulated. b Venn diagram showing the overall differentially expressed TFs. c The number of TF of different families identified in the DEGs of three comparisons. (PPTX 189 kb)
11033_2021_7104_MOESM4_ESM.pptx
Supplementary Fig. S3 a KEGG pathway analysis showing the enrichment of DEGs in “Phenylpropanoid Biosynthesis pathway”. b Heatmap illustrating the expression pattern of DEGs of three comparisons. (PPTX 761 kb)
11033_2021_7104_MOESM5_ESM.pptx
Supplementary Fig. S4 Plant hormone signal transduction pathway showing the expression pattern of three companions. (PPTX 1060 kb)
11033_2021_7104_MOESM6_ESM.pptx
Supplementary Fig. S5 MAPK pathway illustrating the enrichment of DEGs and heatmap showing the expression pattern of all DEGs of three comparisons. (PPTX 974 kb)
Rights and permissions
About this article
Cite this article
Tariq, A., Jabeen, Z., Farrakh, S. et al. Exploring the genetic potential of Pakistani soybean cultivars through RNA-seq based transcriptome analysis. Mol Biol Rep 49, 2889–2897 (2022). https://doi.org/10.1007/s11033-021-07104-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-07104-3