Abstract
Background
The Sugar Will Eventually Be Exported Transporters (SWEET), consisting of the MtN3 and salvia domain, are sugar transporters having an active role in diverse activities in plants such as pollen nutrition, phloem loading, nectar secretion, reproductive tissue development, and plant-pathogen interaction. The SWEET genes have been characterized only in a few fruit crop species.
Methods and results
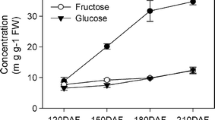
In this study, a total of 15 SWEET genes were identified in the pomegranate (Punica granatum) genome. The gene structure, transmembrane (TM) helices, domain architecture, and phylogenetic relationships of these genes were evaluated using computational approaches. Genes were further classified as Semi-SWEETs or SWEETs based on the TM domains. Similarly, pomegranate, Arabidopsis, rice, and soybean SWEETs were studied together to classify into major groups. In addition, analysis of RNAseq transcriptome data was performed to study SWEEET gene expression dynamics in different tissue. The expression suggests that SWEETs are mostly expressed in pomegranate peel. In addition, PgSWEET13 was found to be differentially expressed under high salinity stress in pomegranate. Further, quantitative PCR analysis confirmed the expression of four candidate genes in leaf and stem tissues.
Conclusion
The information provided here will help to understand the role of SWEET genes in fruit development and under abiotic stress conditions in pomegranate.





Similar content being viewed by others
Data availability
All the data provided as electronic supplementary material along with the article.
References
Patil G, Valliyodan B, Deshmukh R, Prince S, Nicander B, Zhao M, Sonah H, Song L, Lin L, Chaudhary J (2015) Soybean (Glycine max) SWEET gene family: insights through comparative genomics, transcriptome profiling and whole genome re-sequence analysis. BMC Genomics 16:520
Lanjie Z, Jinbo Y, Wei C, Yan L, Youjun L, Yan G, Junyi W, Li Y, Ziyang L, Zhang Y (2018) A genome-wide analysis of SWEET gene family in cotton and their expressions under different stresses. J Cotton Res 1:7
Kühn C, Grof CP (2010) Sucrose transporters of higher plants. Curr Opin Plant Biol 13:287–297
Ayre BG (2011) Membrane-transport systems for sucrose in relation to whole-plant carbon partitioning. Mol Plant 4:377–394
Slewinski TL (2011) Diverse functional roles of monosaccharide transporters and their homologs in vascular plants: a physiological perspective. Mol Plant 4:641–662
Chen L-Q, Hou B-H, Lalonde S, Takanaga H, Hartung ML, Qu X-Q, Guo W-J, Kim J-G, Underwood W, Chaudhuri B (2010) Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468:527
Yuan M, Wang S (2013) Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant 6:665–674
Feng C-Y, Han J-X, Han X-X, Jiang J (2015) Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 573:261–272
Mizuno H, Kasuga S, Kawahigashi H (2016) The sorghum SWEET gene family: stem sucrose accumulation as revealed through transcriptome profiling. Biotechnol Biofuels 9:127
Hu LP, Zhang F, Song SH, Tang XW, Hui XU, Liu GM, Yaqin WA, He HJ (2017) Genome-wide identification, characterization, and expression analysis of the SWEET gene family in cucumber. J Integr Agric 16(7):1486–1501
Gao Y, Wang ZY, Kumar V, Xu XF, Yuan DP, Zhu XF, Li TY, Jia B, Xuan YH (2018) Genome-wide identification of the SWEET gene family in wheat. Gene 642:284–292
Doidy J, Vidal U, Lemoine R (2019) Sugar transporters in Fabaceae, featuring SUT MST and SWEET families of the model plant Medicago truncatula and the agricultural crop Pisum sativum. PLoS ONE 14:e0223173
Xuan YH, Hu YB, Chen L-Q, Sosso D, Ducat DC, Hou B-H, Frommer WB (2013) Functional role of oligomerization for bacterial and plant SWEET sugar transporter family. Proc Natl Acad Sci 110:E3685–E3694
Chen L-Q, Qu X-Q, Hou B-H, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335:207–211
Chardon F, Bedu M, Calenge F, Klemens PA, Spinner L, Clement G, Chietera G, Léran S, Ferrand M, Lacombe B (2013) Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr Biol 23:697–702
Guo W-J, Nagy R, Chen H-Y, Pfrunder S, Yu Y-C, Santelia D, Frommer WB, Martinoia E (2014) SWEET17, a facilitative transporter, mediates fructose transport across the tonoplast of Arabidopsis roots and leaves. Plant Physiol 164:777–789
Klemens PA, Patzke K, Deitmer J, Spinner L, Le Hir R, Bellini C, Bedu M, Chardon F, Krapp A, Neuhaus HE (2013) Overexpression of the vacuolar sugar carrier AtSWEET16 modifies germination, growth, and stress tolerance in Arabidopsis. Plant Physiol 163:1338–1352
Eom J-S, Chen L-Q, Sosso D, Julius BT, Lin I, Qu X-Q, Braun DM, Frommer WB (2015) SWEETs, transporters for intracellular and intercellular sugar translocation. Curr Opin Plant Biol 25:53–62
Guan Y-F, Huang X-Y, Zhu J, Gao J-F, Zhang H-X, Yang Z-N (2008) RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol 147:852–863
Chen LQ (2014) SWEET sugar transporters for phloem transport and pathogen nutrition. New Phytol 201:1150–1155
Denancé N, Szurek B, Noël LD (2014) Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol 55:469–474
Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B (2013) Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol 200:808–819
Zaka A, Grande G, Coronejo T, Quibod IL, Chen C-W, Chang S-J, Szurek B, Arif M, Cruz CV, Oliva R (2018) Natural variations in the promoter of OsSWEET13 and OsSWEET14 expand the range of resistance against Xanthomonas oryzae pv. oryzae. PLoS ONE 13:e0203711
Sonnewald U (2011) SWEETS–the missing sugar efflux carriers. Front Plant Sci 2:7
Lin IW, Sosso D, Chen L-Q, Gase K, Kim S-G, Kessler D, Klinkenberg PM, Gorder MK, Hou B-H, Qu X-Q (2014) Nectar secretion requires sucrose phosphate synthases and the sugar transporter SWEET9. Nature 508:546
Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, Eom J-S, Li C, Nguyen H, Liu B (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37:1344–1350
Vats S, Kumawat S, Kumar V, Patil GB, Joshi T, Sonah H, Sharma TR, Deshmukh R (2019) Genome editing in plants: exploration of technological advancements and challenges. Cells 8:1386
Varshney RK, Godwin ID, Mohapatra T, Jones JD, McCouch SR (2019) A SWEET solution to rice blight. Nat Biotechnol 37:1280–1282
Ben-Simhon Z, Judeinstein S, Nadler-Hassar T, Trainin T, Bar-Ya’akov I, Borochov-Neori H, Holland D (2011) A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 234:865–881
Yuan Z, Fang Y, Zhang T, Fei Z, Han F, Liu C, Liu M, Xiao W, Zhang W, Wu S (2018) The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol J 16:1363–1374
Luo X, Li H, Wu Z, Yao W, Zhao P, Cao D, Yu H, Li K, Poudel K, Zhao D (2019) The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft-and hard-seeded cultivars. Plant Biotechnol J 18:955
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic acids symposium series. Information Retrieval Ltd, London, pp 95–98
El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A (2018) The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Hu B, Jin J, Guo A-Y, Zhang H, Luo J, Gao G (2014) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297
Arvidsson S, Kwasniewski M, Riaño-Pachón DM, Mueller-Roeber B (2008) QuantPrime–a flexible tool for reliable high-throughput primer design for quantitative PCR. BMC Bioinformatics 9:465
Sellami S, Le Hir R, Thorpe MR, Vilaine F, Wolff N, Brini F, Dinant S (2019) Salinity effects on sugar homeostasis and vascular anatomy in the stem of the Arabidopsis thaliana inflorescence. Int J Mol Sci 20:3167
Flagel LE, Wendel JF (2009) Gene duplication and evolutionary novelty in plants. New Phytol 183:557–564
Qin G, Xu C, Ming R, Tang H, Guyot R, Kramer EM, Hu Y, Yi X, Qi Y, Xu X (2017) The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J 91:1108–1128
Zheng Q-M, Tang Z, Xu Q, Deng X-X (2014) Isolation, phylogenetic relationship and expression profiling of sugar transporter genes in sweet orange (Citrus sinensis). Plant Cell Tissue Org Cult (PCTOC) 119:609–624
Jia B, Zhu XF, Pu ZJ, Duan YX, Hao LJ, Zhang J, Chen L-Q, Jeon CO, Xuan YH (2017) Integrative view of the diversity and evolution of SWEET and SemiSWEET sugar transporters. Front Plant Sci 8:2178
Anjali A, Fatima U, Manu M, Ramasamy S, Senthil-Kumar M (2020) Structure and regulation of SWEET transporters in plants: an update. Plant Physiol Biochem
Zhang R, Niu K, Ma H (2020) Identification and expression analysis of the SWEET gene family from Poa pratensis under abiotic stresses. DNA Cell Biol 39:1606
Baker RF, Leach KA, Braun DM (2012) SWEET as sugar: new sucrose effluxers in plants. Mol Plant 5:766–768
Acknowledgements
Authors are thankful to the Department of Biotechnology (DBT), India for the Ramalingaswami fellowship to HS; Council of Scientific and Industrial Research (CSIR) for Shyama Prasad Mukherjee Fellowship (SPMF) to SV, and Junior Research Fellowship (JRF) to YS and VK; University Grants Commission (UGC) for JRF to SK, SS, and GR. The authors are also thankful to Dr. SM Shivaraj for his help in English improvement and valuable suggestions.
Funding
Department of Biotechnology (DBT), India; Science and Engineering Research Board (SERB), Department of Science and Technology, Government of India.
Author information
Authors and Affiliations
Contributions
SK performed qPCR analysis and transcriptome profiling, wrote the first draft of the MS; YS, SV, SS, SS, and RM, contributed in drafting and data analysis, GR, VK, and NR, performed computational analysis, HS conceptualize the study, supervised and finalized the draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2021_6961_MOESM1_ESM.tif
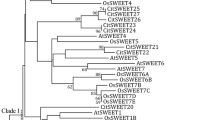
Supplementary file1 (TIF 26384 kb) Supplementary Figure 1 Phylogenetic tree of SWEET genes identified in Punica granatum. The clade I, clade II, clade III, and clade IV is marked in red, purple, blue, and green, respectively. Pomegranate SWEETs is abbreviated as Pg.
11033_2021_6961_MOESM2_ESM.tif
Supplementary file2 (TIF 26390 kb) Supplementary Figure 2 Conserved motifs in the protein sequences of 15 SWEETs identified in the pomegranate genome (PgSWEETs). The conserved motifs were identified by the MEME server (http://meme-suite.org/).
11033_2021_6961_MOESM4_ESM.xlsx
Supplementary file4 (XLSX 9 kb) Supplementary Table 2 Details of Blastp results performed using BioEdit showing highest bit score against respective queries from Arabidopsis thaliana, Glycine max, and Oryza sativa
11033_2021_6961_MOESM5_ESM.xlsx
Supplementary file5 (XLSX 9 kb) Supplementary Table 3 Conserved domain analysis of identified Pomegranate SWEET using NCBI conserved Domain Search
11033_2021_6961_MOESM6_ESM.xlsx
Supplementary file6 (XLSX 10 kb) Supplementary Table 4 Biosequence analysis of Pomegranate SWEET using profile hidden Markov Models (HMMER) (https://www.ebi.ac.uk/Tools/hmmer/)
11033_2021_6961_MOESM7_ESM.xlsx
Supplementary file7 (XLSX 9 kb) Supplementary Table 5 Transmembrane domain identified by using TMHMM and SOSUI servers in Pomegranate SWEET
Rights and permissions
About this article
Cite this article
Kumawat, S., Sharma, Y., Vats, S. et al. Understanding the role of SWEET genes in fruit development and abiotic stress in pomegranate (Punica granatum L.). Mol Biol Rep 49, 1329–1339 (2022). https://doi.org/10.1007/s11033-021-06961-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06961-2