Abstract
Background
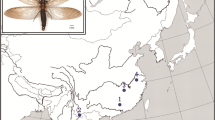
We used Illumina paired-end sequencing to isolate and characterize microsatellites of Canthon cyanellus, a Neotropical roller dung beetle, encompassing several lineages within its distribution range.
Methods and results
We examined C. cyanellus specimens collected at eight different localities in Mexico (two or three specimens per locality). We initially performed amplification tests with 16 loci, but two of which were unsuccessful. The 14 remaining microsatellites were polymorphic, with 2–16 alleles each. The expected and observed heterozygosity ranged from 0.11 to 0.76 and from 0.20 to 0.78, respectively.
Conclusions
These microsatellites will help to assess structure at the population and lineage levels, identify zones of potential hybridization between lineages, and draw a more precise geographic delimitation of C. cyanellus lineages.


Similar content being viewed by others
Data availability
The database of microsatellite genotypes created and analyzed in this study is included in the Supplementary Material 1.xlsx file that accompanies the published article. Additional data supporting the findings of this study are available, upon request, from the corresponding author.
Code availability
Not applicable.
References
Favila ME, Díaz A (1996) Canthon cyanellus cyanellus LeConte (Coleoptera: Scarabaeidae) makes a nest in the field with several brood balls. Coleopt Bull 50:52–60
Favila ME (2001) Historia de vida y comportamiento de un escarabajo necrófago: Canthon cyanellus cyanellus LeConte (coleoptera: Scarabaeinae). Folia Entomológica Mex 40:245–278
Arellano L, León-Cortés JL, Ovaskainen O (2008) Patterns of abundance and movement in relation to landscape structure: a study of a common scarab (Canthon cyanellus cyanellus) in Southern Mexico. Landsc Ecol 23:69–78. https://doi.org/10.1007/s10980-007-9165-8
Portela Salomão R, González-Tokman D, Dáttilo W et al (2018) Landscape structure and composition define the body condition of dung beetles (Coleoptera: Scarabaeinae) in a fragmented tropical rainforest. Ecol Indic 88:144–151. https://doi.org/10.1016/j.ecolind.2018.01.033
Robinson M (1948) A review of the species of Canthon inhabiting the United States (Scarabaeidae: Coleoptera). Trans Am Entomol Soc 1890–74:83–100
Solís-Blanco A, Kohlmann B (2002) El género Canthon (Coleoptera: Scarabaeidae) en Costa Rica. G Ital Entomol 10:1–68
Halffter-Salas G (1961) Monografía de las especies norteamericanas del género Canthon Hoffsg. (Coleoptera: Scarabaeidae). Univ Zulia 20:225–320
Blackwelder RE (1944) Checklist of the coleopterous insects of Mexico, Central America, the West Indies, and South America. US Government Printing Office, Washington
Bellés X, Favila M (1983) Protection chimique du nid chez Canthon cyanellus cyanellus LeConte [Col. Scarabaeidae]. Bull Société Entomol Fr 88:602–607
Cortez V, Favila ME, Verdú JR, Ortiz AJ (2012) Behavioral and antennal electrophysiological responses of a predator ant to the pygidial gland secretions of two species of neotropical dung roller beetles. Chemoecology 22:29–38
Cortez V, Verdú JR, Ortiz AJ et al (2015) Chemical diversity and potential biological functions of the pygidial gland secretions in two species of Neotropical dung roller beetles. Chemoecology 25:201–213. https://doi.org/10.1007/s00049-015-0189-2
Favila M, Chamorro-Florescano I (2008) Male reproductive status affects contest outcome during nidification in Canthon cyanellus cyanellus LeConte (Coleoptera: scarabaeidae). Behaviour 145:1811–1821. https://doi.org/10.1163/156853908786279637
Favila M, Chamorro-Florescano I (2009) The reproductive status of both sexes affects the frequency of mating and the reproductive success of males in the ball roller beetle Canthon cyanellus cyanellus (coleoptera: Scarabaeidae). Behaviour 146:1499–1512. https://doi.org/10.1163/156853909X445560
Chamorro-Florescano IA, Favila ME, Macías-Ordóñez R (2011) Ownership, size and reproductive status affect the outcome of food ball contests in a dung roller beetle: when do enemies share? Evol Ecol 25:277–289
Chamorro-Florescano IA, Favila ME (2016) Male success in intrasexual contests extends to the level of sperm competition in a species of dung roller beetle. Ethology 122:53–60. https://doi.org/10.1111/eth.12443
Chamorro-Florescano IA, Favila ME, Macías-Ordóñez R (2017) Contests over reproductive resources in female roller beetles: outcome predictors and sharing as an option. PLoS ONE 12:e0182931. https://doi.org/10.1371/journal.pone.0182931
Favila ME, Ortiz-Domínguez M, Chamorro-Florescano I, Cortez-Gallardo V (2012) Comunicación química y comportamiento reproductor de los escarabajos rodadores del estiércol (Scarabaeinae: Scarabaeini): aspectos ecológicos y evolutivos, y sus posibles aplicaciones [pp. 141–164]. Temas Sel En Ecol Quím Insectos JC Rojas EA Malo Ed El Col Front Sur Tapachula Mex
Favila ME, Nolasco J, Florescano IC, Equihua M (2005) Sperm competition and evidence of sperm fertilization patterns in the carrion ball-roller beetle Canthon cyanellus cyanellus LeConte (Scarabaeidae: Scarabaeinae). Behav Ecol Sociobiol 59:38. https://doi.org/10.1007/s00265-005-0006-y
Ortiz-Domínguez M, Favila ME, Mendoza-López MR et al (2006) Epicuticular compounds and sexual recognition in the ball-roller scarab, Canthon cyanellus cyanellus. Entomol Exp Appl 119:23–27. https://doi.org/10.1111/j.1570-7458.2006.00388.x
Ortíz-Domínguez M, Favila ME, Mendoza-López MR (2006) Mate recognition differences among allopatric populations of the scarab Canthon cyanellus cyanellus (Coleoptera: Scarabaeidae). Ann Entomol Soc Am 99:1248–1256. https://doi.org/10.1603/0013-8746(2006)99[1248:MRDAAP]2.0.CO;2
Ortiz-Domínguez M, Favila-Castillo ME, González D, Zuñiga D (2010) Genetic differences in populations of the ball roller scarab Canthon cyanellus cyanellus (coleoptera: Scarabaeidae): a preliminary analysis. Elytron 24:99–106
Chung H, Carroll SB (2015) Wax, sex and the origin of species: dual roles of insect cuticular hydrocarbons in adaptation and mating. BioEssays 37:822–830. https://doi.org/10.1002/bies.201500014
Nolasco-Soto J, González-Astorga J, Espinosa Monteros A et al (2017) Phylogeographic structure of Canthon cyanellus (coleoptera: Scarabaeidae), a Neotropical dung beetle in the Mexican transition Zone: insights on its origin and the impacts of pleistocene climatic fluctuations on population dynamics. Mol Phylogenet Evol 109:180–190. https://doi.org/10.1016/j.ympev.2017.01.004
Nolasco-Soto J, Favila ME, De Los E, Monteros A et al (2020) Variations in genetic structure and male genitalia suggest recent lineage diversification in the neotropical dung beetle complex Canthon cyanellus (Scarabaeidae: Scarabaeinae). Biol J Linn Soc 131:505–520. https://doi.org/10.1093/biolinnean/blaa131
Ratnasingham S, Hebert PDN (2013) A DNA-based registry for all animal species: the barcode index number (BIN) system. PLoS ONE 8:e66213. https://doi.org/10.1371/journal.pone.0066213
Vieira MLC, Santini L, Diniz AL et al (2016) Microsatellite markers: what they mean and why they are so useful. Genet Mol Biol 39:312–328. https://doi.org/10.1590/1678-4685-GMB-2016-0027
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics Babraham Institute, Cambridge, United Kingdom
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17:10–12. https://doi.org/10.14806/ej.17.1.200
Bankevich A, Nurk S, Antipov D et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. https://doi.org/10.1089/cmb.2012.0021
Meglécz E, Pech N, Gilles A et al (2014) QDD version 3.1: a user-friendly computer program for microsatellite selection and primer design revisited: experimental validation of variables determining genotyping success rate. Mol Ecol Resour 14:1302–1313. https://doi.org/10.1111/1755-0998.12271
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234. https://doi.org/10.1038/72708
Guichoux E, Lagache L, Wagner S et al (2011) Current trends in microsatellite genotyping. Mol Ecol Resour 11:591–611. https://doi.org/10.1111/j.1755-0998.2011.03014.x
Peakall R, Smouse PE (2006) genalex 6: genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Liu K, Muse SV (2005) PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129. https://doi.org/10.1093/bioinformatics/bti282
Valière N (2002) gimlet: a computer program for analysing genetic individual identification data. Mol Ecol Notes 2:377–379. https://doi.org/10.1046/j.1471-8286.2002.00228.x-i2
Amos W, Hoffman JI, Frodsham A et al (2007) Automated binning of microsatellite alleles: problems and solutions. Mol Ecol Notes 7:10–14. https://doi.org/10.1111/j.1471-8286.2006.01560.x
Li CC (1976) First course in population genetics. Boxwood, Pac Grove CA
Manjeri G, Muhamad R, Faridah QZ, Tan SG (2014) Development of single locus DNA microsatellite markers in Oryctes rhinoceros (Linnaeus) using 5′ anchored RAMs-PCR method. J Genet 93:92–96. https://doi.org/10.1007/s12041-012-0189-8
Behura SK, Severson DW (2015) Motif mismatches in microsatellites: insights from genome-wide investigation among 20 insect species. DNA Res 22:29–38. https://doi.org/10.1093/dnares/dsu036
Song X, Yang T, Yan X et al (2020) Comparison of microsatellite distribution patterns in twenty-nine beetle genomes. Gene 757:144919. https://doi.org/10.1016/j.gene.2020.144919
Santana QC, Coetzee MPA, Steenkamp ET et al (2009) Microsatellite discovery by deep sequencing of enriched genomic libraries. Biotechniques 46:217–223. https://doi.org/10.2144/000113085
Katti MV, Ranjekar PK, Gupta VS (2001) Differential distribution of simple sequence repeats in eukaryotic genome sequences. Mol Biol Evol 18:1161–1167. https://doi.org/10.1093/oxfordjournals.molbev.a003903
Tóth G, Gáspári Z, Jurka J (2000) Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res 10:967–981. https://doi.org/10.1101/gr.10.7.967
Eder MLR, Rosa AL (2020) Non-tandem repeat polymorphisms at microsatellite loci in wine yeast species. Mol Genet Genomics 295:685–693. https://doi.org/10.1007/s00438-020-01652-2
Sampaio P, Correia A, Gusmão L et al (2007) Sequence analysis reveals complex mutational processes for allele length variation at two polymorphic microsatellite loci in Candida albicans. Communicating current research and educational topics and trends in applied microbiology. Formatex, Badajoz, pp 926–935
Angers B, Bernatchez L (1997) Complex evolution of a salmonid microsatellite locus and its consequences in inferring allelic divergence from size information. Mol Biol Evol 14:230–238. https://doi.org/10.1093/oxfordjournals.molbev.a025759
Schlötterer C, Zangerl B (1999) The use of imperfect microsatellites for DNA fingerprinting and population genetics. In: Epplen JT, Lubjuhn T (eds) DNA profiling and DNA fingerprinting. Birkhäuser, Basel, pp 153–165
Viard F, Franck P, Dubois M-P et al (1998) Variation of microsatellite size homoplasy across electromorphs, loci, and populations in three invertebrate species. J Mol Evol 47:42–51
Von Wahlund S (1928) Zusammensetzung von population und Korrelationsers cheimungen von Standpunkt der veierbungs lehreaus betachlet. Hereditas 11:65–106
Acknowledgements
Sequencing was performed at the Genomics Core Facility of Servicio Central de Soporte a la Investigación Experimental (SCSIE), Universidad de Valencia. This study was funded by a research grant (CB 282922) from Consejo Nacional de Ciencia y Tecnología (CONACyT) awarded to Rosa Ana Sánchez Guillén. María Elena Sánchez-Salazar edited the English manuscript.
Funding
This study was funded by a research grant from Consejo Nacional de Ciencia y Tecnología (CONACyT) awarded to Rosa Ana Sánchez Guillén (CB 282922).
Author information
Authors and Affiliations
Contributions
All the authors contributed to the conception and design of this study. Samples were collected by JN-S. Wet lab techniques were performed by JN-S and LRA-V. Data analyses were carried out by RAS-G and LRA-V. The manuscript was written and edited equally by all the authors. All the authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest that are relevant to the content of this article.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Neither the article nor portions of it have been previously published elsewhere. The manuscript is not under consideration for publication in any other journal and will not be submitted elsewhere until the Molecular Biology Reports editorial process is completed. All authors consent to the publication of the manuscript in Molecular Biology Reports should the article be accepted.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11033_2021_6761_MOESM3_ESM.pdf
Supplementary file3 (PDF 362 kb) Sequence, primers, and issues found with the unsuccessfully genotyped microsatellite loci Ccy15 and Ccy16
11033_2021_6761_MOESM4_ESM.tiff
Supplementary file4 (TIFF 201 kb) Binning of the Ccy09 and Ccy12 alleles. Solid colors denote raw fragment lengths binned together into a single allele class
Rights and permissions
About this article
Cite this article
Arce-Valdés, L.R., Sánchez-Guillén, R.A., Nolasco-Soto, J. et al. Next-generation sequencing, isolation and characterization of 14 microsatellite loci of Canthon cyanellus (Coleoptera: Scarabaeidae). Mol Biol Rep 48, 7433–7441 (2021). https://doi.org/10.1007/s11033-021-06761-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06761-8