Abstract
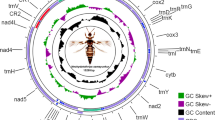
The species Eudocima salaminia (Cramer, 1777) commonly known as the fruit-piercing moth belongs to family Erebidae. Its distribution varies from India and across South-east Asia, pacific islands and parts of Australia. The insect is a devastating pest of citrus, longans and lychees. In the present study, complete mitochondrial genome of Eudocima salaminia was sequenced and analyzed using Illumina sequencer. The phylogenetic tree was reconstructed based on nucleotide sequences of 13 PCGs using Maximum likelihood method-General Reversible mitochondrial (mtREV) model. The mitogenome has 15,597 base pairs (bp) in length, comprising of 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes and A + T-rich region. All protein-coding genes (PCGs) initiate with canonical start codon ATN. The gene order (trnQ-trnI-trnM) of tRNA shows a different rearrangement compared to ancestral insect gene order (trnI-trnQ-trnM). Almost all tRNAs have a typical cloverleaf secondary structure except for trnS1 (AGN) which lacks the dihydrouridine arm. At the beginning of the control region, we observed a conserved polyT”, motif “ATTTA” and microsatellite (TA)n element. There are 21 intergenic regions and five overlapping regions ranging from 1 to 73 bp and 1 to 8 bp, respectively. The phylogenetic relationships based on nucleotide sequences of 13 PCGs using Maximum likelihood method showed the family level relationships as (Notodontidae + (Euteliidae + Noctuidae + (Erebidae + Nolidae))). The present study represents the similarity to phylogenetic analysis of Noctuoidea mitogenome. Moreover, the family Erebidae is the sister to the families of (Euteliidae + Noctuidae + Nolidae).






Similar content being viewed by others
References
Avise JC, Arnold J, Ball RM, Bermingham E, Lamb T, Neigel JE, Reeb CA, Saunders NC (1987) Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. J Mol Evol 18:489. https://doi.org/10.1146/annurev.es.18.110187.002421
Ballard JWO (2000) Comparative genomics of mitochondrial DNA in Drosophila simulans. J Mol Evol 51:64. https://doi.org/10.1007/s002390010067
Ballard JWO, Rand DM (2005) The population biology of mitochondrial DNA and its phylogenetic implications. Annu Rev Ecol Syst 36:621. https://www.jstor.org/stable/30033819
Cameron SL, Whiting MF (2008) The complete mitochondrial genome of the tobacco hornworm, Manduca sexta, (Insecta: Lepidoptera: Sphingidae), and an examination of mitochondrial gene variability within butterflies and moths. Gene 408:112. https://doi.org/10.1016/j.gene.2007.10.023
Hao J, Sun Q, Zhao H, Sun X, Gai Y, Yang Q (2012) The complete mitochondrial genome of Ctenoptilum vasava (Lepidoptera: Hesperiidae: Pyrginae) and its phylogenetic implication. Comp Funct Genomics. https://doi.org/10.1155/2012/328049
Cameron SL (2014) Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol 59:95. https://doi.org/10.1146/annurev-ento-011613-162007
Gissi C, Iannelli F, Pesole G (2008) Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. J Hered 101:301. https://doi.org/10.1038/hdy.2008.62
Liu Q-N, Xin Z-Z, Bian D-D, Chai X-Y, Zhou C-L, Tang B-P (2016) The first complete mitochondrial genome for the subfamily Limacodidae and implications for the higher phylogeny of Lepidoptera. Sci Rep 6:35878. https://doi.org/10.1038/srep35878
Wolstenholme DR (1992) Animal mitochondrial DNA: structure and evolution. Int Rev Cytol 141:173–216. https://doi.org/10.1016/S0074-7696(08)62066-5
Boore JL (1999) Animal mitochondrial genomes. Nucleic Acids Res 27:1767. https://doi.org/10.1093/nar/27.8.1767
Nardi F, Carapelli A, Dallai R, Frati F (2003) The mitochondrial genome of the olive fly Bactrocera oleae: two haplotypes from distant geographical locations. Insect Mol Biol 12:605. https://doi.org/10.1046/j.1365-2583.2003.00445.x
Van Nieukerken EJ, Kaila L, Kitching IJ, Kristensen NP, Lees DC, Minet J, Mitter C, Mutanen M, Regier JC, Simonsen TJ (2011) Order Lepidoptera Linnaeus 1758. In: Zhang Z-Q (ed) Animal biodiversity: an outline of higher-level classification and survey of taxonomic richness, vol 3148. Zootaxa, Waco, p 212. https://doi.org/10.11646/zootaxa.3148.1.41
Kitching IJ (1984) An historical review of the higher classification of the Noctuidae (Lepidoptera). Bull Br Mus (Nat Hist) (Entomol) 49:153
Miller JS (1991) Cladistics and classification of the Notodontidae (Lepidoptera, Noctuoidea) based on larval and adult morphology. Bull AMNH 204:1. http://hdl.handle.net/2246/897
Kitching IJR, Rawlins JE (1998) The Noctuoidea. In: Kristensen NP (ed) Lepidoptera, moths and butterflies. Evolution, systematics and biogeography. Handbook of Zoology, vol 1. Pemberley Books, Iver, p 355
Mitchell A, Mitter C, Regier JC (2006) Systematics and evolution of the cutworm moths (Lepidoptera: Noctuidae): evidence from two protein-coding nuclear genes. Syst Entomol 31:21. https://doi.org/10.1111/j.1365-3113.2005.00306.x
Fang QQ, Mitchell A, Regier JC, Mitter C, Friedlander TP, Poole RW (2000) Phylogenetic utility of the nuclear gene dopa decarboxylase in noctuoid moths (Insecta: Lepidoptera: Noctuoidea). Mol Phylogenet Evol 15:473. https://doi.org/10.1006/mpev.1999.0758
Mitchell ACS, Regier JC, Mitter C, Poole RW, Matthews M (1997) Phylogenetic utility of elongation factor-1 alpha in Noctuoidea (Insecta: Lepidoptera): the limits of synonymous substitution. Mol Biol Evol. https://doi.org/10.1093/oxfordjournals.molbev.a025774
Mitchell A, Mitter C, Regier JC (2000) More taxa or more characters revisited: combining data from nuclear protein-encoding genes for phylogenetic analyses of Noctuoidea (Insecta: Lepidoptera). Syst Biol 49:202. https://doi.org/10.1093/sysbio/49.2.202
Weller SJ, Pashley D, Martin J, Constable J (1994) Phylogeny of noctuoid moths and the utility of combining independent nuclear and mitochondrial genes. Syst Biol 43:194. https://doi.org/10.1093/sysbio/43.2.194
Zahiri R, Kitching IJ, Lafontaine JD, Mutanen M, Kaila L, Holloway JD, Wahlberg N (2011) A new molecular phylogeny offers hope for a stable family level classification of the Noctuoidea (Lepidoptera). Zool Scr 40:158. https://doi.org/10.1111/j.1463-6409.2010.00459.x
Zahiri R, Holloway JD, Kitching IJ, Lafontaine JD, Mutanen M, Wahlberg N (2012) Molecular phylogenetics of Erebidae (Lepidoptera, Noctuoidea). Syst Entomol 37:102. https://doi.org/10.1111/j.1365-3113.2011.00607.x
Liao F, Wang L, Wu S, Li Y-P, Zhao L, Huang G-M, Niu C-J, Liu Y-Q, Li M-G (2010) The complete mitochondrial genome of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae). Int J Biol Sci 6:172. https://doi.org/10.7150/ijbs.6.172
Lu H-F, Su T-J, Luo A-R, Zhu C-D, Wu C-S (2013) Characterization of the complete mitochondrion genome of diurnal moth Amata emma (Butler) (Lepidoptera: Erebidae) and its phylogenetic implications. PLoS ONE 8:e72410. https://doi.org/10.1371/journal.pone.0072410
Sivasankaran K, Mathew P, Anand S, Ceasar SA, Mariapackiam S, Ignacimuthu S (2017) Complete mitochondrial genome sequence of fruit-piercing moth Eudocima phalonia (Linnaeus, 1763) (Lepidoptera: Noctuoidea). Genomics Data 14:66. https://doi.org/10.1016/j.gdata.2017.09.004
Tang M, Tan M, Meng G, Yang S, Su X, Liu S, Song W, Li Y, Wu Q, Zhang A (2014) Multiplex sequencing of pooled mitochondrial genomes—a crucial step toward biodiversity analysis using mito-metagenomics. Nucleic Acids Res 42:e166. https://doi.org/10.1093/nar/gku917
Yang X, Cameron SL, Lees DC, Xue D, Han H (2015) A mitochondrial genome phylogeny of owlet moths (Lepidoptera: Noctuoidea), and examination of the utility of mitochondrial genomes for lepidopteran phylogenetics. Mol Phylogenet Evol 85:230. https://doi.org/10.1016/j.ympev.2015.02.005
Yajun Z, Guoliang Z, Rui F, Jun Y, Jianping Y (2010) The complete sequence determination and analysis of Lymantria dispar (Lepidoptera: Lymantriidae) mitochondrial genome. Plant Quar 24(4):6–11
Yuan M-L, Zhang Q-L (2013) The complete mitochondrial genome of Gynaephora menyuanensis (Lepidoptera: Lymantriidae) from the Qinghai-Tibetan Plateau. Mitochondrial DNA 24:328. https://doi.org/10.3109/19401736.2012.760077
Chai HN, Du YZ (2012) The complete mitochondrial genome of the pink stem borer, Sesamia inferens, in comparison with four other noctuid moths. Int J Mol Sci 13:10236. https://doi.org/10.3390/ijms130810236
Gong Y-J, Wu Q-L, Wei S-J (2013) Complete mitogenome of the Argyrogramma agnata (Lepidoptera: Noctuidae). Mitochondrial DNA 24:391. https://doi.org/10.3109/19401736.2013.763241
Liu Q-N, Bian D-D, Jiang S-H, Ge B-M, Zhou C-L, Tang B-P (2015) Characterization of the complete mitochondrial genome of the oriental armyworm, Mythimna separata (Lepidoptera: Noctuidae). Eur J Entomol 112:399. https://doi.org/10.14411/eje.2015.055
Liu QN, Zhu BJ, Dai LS, Wang L, Qian C, Wei GQ, Liu CL (2014) The complete mitochondrial genome of the common cutworm, Spodoptera litura (Lepidoptera: Noctuidae). Mitochondrial DNA. https://doi.org/10.3109/19401736.2013.873934
Timmermans MJ, Lees DC, Simonsen TJ (2014) Towards a mitogenomic phylogeny of Lepidoptera. Mol Phylogenet Evol 79:169. https://doi.org/10.1016/j.ympev.2014.05.031
Walsh TK (2016) Characterization of the complete mitochondrial genome of the Australian Heliothine moth, Australothis rubrescens (Lepidoptera: Noctuidae). Mitochondrial DNA Part A 27:167. https://doi.org/10.3109/19401736.2013.878927
Wu Q-L, Cui W-X, Du B-Z, Gu Y, Wei S-J (2014) The complete mitogenome of the turnip moth Agrotis segetum (Lepidoptera: Noctuidae). Mitochondrial DNA 25(5):345–347. https://doi.org/10.3109/19401736.2013.800496
Wu Q-L, Cui W-X, Wei S-J (2015) Characterization of the complete mitochondrial genome of the black cutworm Agrotis ipsilon (Lepidoptera: Noctuidae). Mitochondrial DNA 26:139. https://doi.org/10.3109/19401736.2013.815175
Wu Q-L, Gong Y-J, Gu Y, Wei S-J (2013) The complete mitochondrial genome of the beet armyworm Spodoptera exigua (Hübner) (Lepodiptera: Noctuidae). Mitochondrial DNA 24:31. https://doi.org/10.3109/19401736.2012.716052
Yin J, Hong G-Y, Wang A-M, Cao Y-Z, Wei Z-J (2010) Mitochondrial genome of the cotton bollworm Helicoverpa armigera (Lepidoptera: Noctuidae) and comparison with other Lepidopterans. Mitochondrial DNA 21:160. https://doi.org/10.3109/19401736.2010.503242
Holloway JD (2005) The moths of Borneo: family Noctuidae, subfamily Catocalinae. Malay Nat J 58:1–529
Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181. https://doi.org/10.1093/nar/gkn179
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547. https://doi.org/10.1093/molbev/msy096
Liu Q-N, Chai X-Y, Bian D-D, Ge B-M, Zhou C-L, Tang B-P (2016) The complete mitochondrial genome of fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae). Genes Genomics 38:205. https://doi.org/10.1007/s13258-015-0346-6
Wu Q-L, Li Q, Gu Y, Shi B-C, van Achterberg C, Wei S-J, Chen X-X (2014) The complete mitochondrial genome of Taeniogonalos taihorina (Bischoff) (Hymenoptera: Trigonalyidae) reveals a novel gene rearrangement pattern in the Hymenoptera. Gene 543:76. https://doi.org/10.1016/j.gene.2014.04.003
Djoumad A, Nisole A, Zahiri R, Freschi L, Picq S, Gundersen-Rindal DE, Sparks ME, Dewar K, Stewart D, Maaroufi H (2017) Comparative analysis of mitochondrial genomes of geographic variants of the gypsy moth, Lymantria dispar, reveals a previously undescribed genotypic entity. Sci Rep 7:1. https://doi.org/10.1038/s41598-017-14530-6
Dowton M, Castro LR, Campbell SL, Bargon SD, Austin AD (2003) Frequent mitochondrial gene rearrangements at the hymenopteran nad3–nad5 junction. J Mol Evol 56:517. https://doi.org/10.1007/s00239-002-2420-3
Clary DO, Wolstenholme DR (1985) The mitochondrial DNA molecule of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol 22:252. https://doi.org/10.1007/BF02099755
San Mauro D, Gower DJ, Zardoya R, Wilkinson M (2006) A hotspot of gene order rearrangement by tandem duplication and random loss in the vertebrate mitochondrial genome. Mol Biol Evol 23:227. https://doi.org/10.1093/molbev/msj025
Jiang S-T, Hong G-Y, Yu M, Li N, Yang Y, Liu Y-Q, Wei Z-J (2009) Characterization of the complete mitochondrial genome of the giant silkworm moth, Eriogyna pyretorum (Lepidoptera: Saturniidae). Int J Biol Sci 5:351. https://doi.org/10.7150/ijbs.5.351
Liu Q-N, Zhu B-J, Dai L-S, Liu C-L (2013) The complete mitogenome of Bombyx mori strain Dazao (Lepidoptera: Bombycidae) and comparison with other lepidopteran insects. Genomics 101:64. https://doi.org/10.1016/j.ygeno.2012.10.002
Yang X, Xue D, Han H (2013) The complete mitochondrial genome of Biston panterinaria (Lepidoptera: Geometridae), with phylogenetic utility of mitochondrial genome in the Lepidoptera. Gene 515:349. https://doi.org/10.1016/j.gene.2012.11.031
Dai LQC, Zhang C, Wang L, Wei G, Li J (2015) Characterization of the complete mitochondrial genome of Cerura menciana and comparison with other Lepidopteran insects. PLoS ONE 10(8):0132951. https://doi.org/10.1371/journal.pone.0132951
Liu Y, Xin Z-Z, Zhu X-Y, Zhao X-M, Wang Y, Tang B-P, Zhang H-B, Zhang D-Z, Zhou C-L, Liu Q-N (2017) The complete mitochondrial genome of Euproctis similis (Lepidoptera: Noctuoidea: Erebidae) and phylogenetic analysis. Int J Biol Macromol 105:219. https://doi.org/10.1016/j.ijbiomac.2017.07.033
Zhu X-Y, Xin Z-Z, Liu Y, Wang Y, Huang Y, Yang Z-H, Chu X-H, Zhang D-Z, Zhang H-B, Zhou C-L (2018) The complete mitochondrial genome of Clostera anastomosis (Lepidoptera: Notodontidae) and implication for the phylogenetic relationships of Noctuoidea species. Int J Biol Macromol 118:1574. https://doi.org/10.1016/j.ijbiomac.2018.06.188
Yang M, Song L, Shi Y, Yin Y, Wang Y, Zhang P, Chen J, Lou L, Liu X (2019) The complete mitochondrial genome of a medicinal insect, Hydrillodes repugnalis (Lepidoptera: Noctuoidea: Erebidae), and related phylogenetic analysis. Int J Biol Macromol 123:485. https://doi.org/10.1016/j.ijbiomac.2018.10.149
Xin Z-Z, Liu Y, Zhang D-Z, Wang Z-F, Tang B-P, Zhang H-B, Zhou C-L, Chai X-Y, Liu Q-N (2018) Comparative mitochondrial genome analysis of Spilarctia subcarnea and other noctuid insects. Int J Biol Macromol 107:121. https://doi.org/10.1016/j.ijbiomac.2017.08.153
Huang Y, Liu Y, Zhu X-Y, Xin Z-Z, Zhang H-B, Zhang D-Z, Wang J-L, Tang B-P, Zhou C-L, Liu Q-N (2019) Comparative mitochondrial genome analysis of Grammodes geometrica and other noctuid insects reveals conserved mitochondrial genome organization and phylogeny. Int J Biol Macromol 125:1257. https://doi.org/10.1016/j.ijbiomac.2018.09.104
Sun Y, Zhu Y, Chen C, Zhu Q, Zhu Q, Zhou Y, Zhou X, Zhu P, Li J, Zhang H (2020) The complete mitochondrial genome of Dysgonia stuposa (Lepidoptera: Erebidae) and phylogenetic relationships within Noctuoidea. PeerJ 8:e8780. https://doi.org/10.7717/peerj.8780
Dai L-S, Zhu B-J, Zhao Y, Zhang C-F, Liu C-L (2016) Comparative mitochondrial genome analysis of Eligma narcissus and other Lepidopteran insects reveals conserved mitochondrial genome organization and phylogenetic relationships. Sci Rep 6:1. https://doi.org/10.1038/srep26387
Sun Q, Sun X, Wang X, Gai Y, Hu J, Zhu C, Hao J (2012) Complete sequence of the mitochondrial genome of the Japanese buff-tip moth, Phalera flavescens (Lepidoptera: Notodontidae). Genet Mol Res 11:4213–4225. https://doi.org/10.4238/2012
Zhang D-X, Hewitt GM (1997) Insect mitochondrial control region: a review of its structure, evolution and usefulness in evolutionary studies. Biochem Syst Ecol 25:99. https://doi.org/10.1016/S0305-1978(96)00042-7
Li J, Zhao Y, Lin R, Zhang Y, Hu K, Li Y, Huang Z, Peng S, Ding J, Geng X (2019) Mitochondrial genome characteristics of Somena scintillans (Lepidoptera: Erebidae) and comparation with other Noctuoidea insects. Genomics 111:1239. https://doi.org/10.1016/j.ygeno.2018.08.003
Acknowledgements
This study was funded by the Department of Science and Technology (DST)-Science and Engineering Research Board (SERB) [Grant Number: EMR/2017/000566]. The authors are grateful to the Director, Entomology Research Institute, Loyola College, Chennai, India for providing the facilities. We also thank Dr Stanislaus Antony Ceasar, Senior Scientist, Department of Bioscience, Rajagiri College of Social Sciences, Rajagiri Valley, Kakkanad, Kochi, Kerala, India for helping in validating the data.
Funding
The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
MR and RAS: Conceived the experiment and data curation. SI: Supervision, reviewing and editing the manuscript. SK: Designed, conceived the experiment, performed the genomic analyses, writing—original draft preparation. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare there are no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Riyaz, M., Shah, R.A., Savarimuthu, I. et al. Comparative mitochondrial genome analysis of Eudocima salaminia (Cramer, 1777) (Lepidoptera: Noctuoidea), novel gene rearrangement and phylogenetic relationship within the superfamily Noctuoidea. Mol Biol Rep 48, 4449–4463 (2021). https://doi.org/10.1007/s11033-021-06465-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06465-z