Abstract
Brassica rapa L., which includes Chinese cabbage, turnip, and pak choi, has more complex flowering time regulation than does Arabidopsis thaliana due to the presence of multiple paralogous flowering time genes. FLOWERING LOCUS C (FLC) is one of the key genes regulating the flowering time, and B. rapa has four FLC paralogs. BrFLC5 on the reference genome is deemed a pseudogene because of a mutation (from G to A) in the splice site of the third intron, but there are some accessions with a G nucleotide in the splice site. In this study, we genotyped 310 B. rapa accessions and found that 19 had homozygous and 81 had heterozygous putative functional BrFLC5 alleles. Accessions of turnip showed the highest proportion with a functional BrFLC5 allele. BrFLC5 acts as a floral repressor when overexpressed in A. thaliana. The BrFLC5 expression level varied in pre-vernalized plants, and this transcriptional variation was not associated with the G/A polymorphism in the third intron. Three accessions having a higher BrFLC5 expression in pre-vernalized plants had a 584-bp insertion in the promoter region. Many regions homologous to this 584-bp sequence are present in the B. rapa genome, and this 584-bp inserted region has tandem duplications of an AT-rich sequence in its central region. The possibility that a high expression of a functional BrFLC5 could contribute to producing premature bolting-resistant lines in B. rapa vegetables is discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Brassica rapa L. comprises a large number of morphological variants and includes commercially important leafy vegetables such as Chinese cabbage (var. pekinensis), pak choi (var. chinensis), komatsuna (var. perviridis), root vegetables such as turnip (var. rapa), and oil seed/mustard (var. trilocularis, var. dichotoma, var. oleifera) (Lv et al. 2020). B. rapa leafy vegetables are grown mainly in Asian countries, and root vegetables are grown in European countries and the USA as well as in Asian countries. In India, oil seeds and mustard are the main crops (Qi et al. 2017). The whole-genome sequence of B. rapa was the first to be determined in the genus Brassica using the Chinese cabbage doubled haploid line, Chiifu-401-42 (Wang et al. 2011), followed by the whole genome sequence in yellow sarson (Belser et al. 2018) and pak choi (Li et al. 2020; Li et al. 2021). The reference genome sequence of Chiifu-401-42 was updated by long-read sequencing technology and high-through chromosome conformation capture (Hi-C) technology (Zhang et al. 2018).
Flowering time is one of the major influences on the quality of the harvested product and seed in the genus Brassica. Brassica species originally required prolonged cold exposure to induce flowering, and this phenomenon is called vernalization (Itabashi et al. 2018; Akter et al. 2018, 2021). B. rapa has two main flowering types; one requires little or no vernalization, and the other requires vernalization (Su et al. 2018). Even within B. rapa accessions requiring vernalization, there is a diversity in the degree of vernalization required, a variation of the length of the period of cold exposure for flowering. For oilseed or mustard, where seed is the product and crops are grown in warm climates, accessions with little or no vernalization requirement are used. Most leafy or root vegetables have a high vernalization requirement because early bolting leads to a decrease in quality of products, especially in spring Chinese cabbage that is sown in late winter or early spring (Akter et al. 2021).
The floral repressor, FLOWERING LOCUS C (FLC), a MADS-box transcription factor, mediates vernalization in B. rapa (Shea et al. 2019; Takada et al. 2019; Akter et al. 2021). Due to the whole genome triplication (WGT) in B. rapa, there are four copies of BrFLC (BrFLC1, BrFLC2, BrFLC3, and BrFLC5) (Shea et al. 2018; Akter et al. 2019, 2021). All four BrFLC paralogs are expressed in pre-vernalized plants and have active histone marks, histone H3 lysine 4 trimethylation (H3K4me3) and H3K36me3, in their coding regions (Mehraj et al. 2021). The expression of all four BrFLC paralogs gradually decreases during prolonged cold exposure (Akter et al. 2020). During this period, the repressive histone mark, H3K27me3, accumulates around the transcription start site of BrFLCs (Akter et al. 2019). After returning to a warm environment, H3K27me3 spreads across the entire encoding regions of all BrFLC paralogs, and the repression of their expression is maintained (Kawanabe et al. 2016; Akter et al. 2019, 2021).
There are variations in expression levels in pre-vernalized plants and in the degree of suppression of expression following vernalization among the four BrFLC paralogs in the same plant (Takada et al. 2019). Furthermore, there are variations in expression levels of each BrFLC gene between accessions in pre-vernalized plants and in the degree of suppression of expression following vernalization in each BrFLC gene between accessions (Takada et al. 2019). The total expression level of the four BrFLC paralogs in pre-vernalized and the degree of suppression of expression in each BrFLC following prolonged cold treatment are important factors for the intraspecific variation of the vernalization requirement (Takada et al. 2019; Akter et al. 2021). These two characteristics also contribute to the difference in vernalization requirements in canola (Brassica napus); winter types, which have high vernalization requirements, showed a high level of total BnFLC expression or residual BnFLC expression level in specific paralogs following prolonged cold treatment (Calderwood et al. 2021). Since the total BrFLC expression level in pre-vernalized plants is important, the loss of function of one BrFLC paralog will result in a reduction in the total expression level of functional BrFLCs and a reduction in the vernalization requirement. For example, the early flowering accession, yellow sarson, has a loss of function of BrFLC1 and BrFLC2 (Li et al. 2009; Yuan et al. 2009). Similarly, if one BrFLC paralog is not reduced in expression following prolonged cold treatment, the repression rate of total BrFLC expression will be reduced, resulting in a higher vernalization requirement. For example, in Tsukena No. 2, the expression levels of BrFLC2 and BrFLC3 are less reduced following prolonged cold treatment (Kitamoto et al. 2014), and the expression level of BrFLC1 is less reduced in BRA2209 (Takada et al. 2019); these two accessions both have a high vernalization requirement.
BrFLC5 is classified as a pseudogene due to the deletion of two exons in the reference genome (Schranz et al. 2002; Wang et al. 2011). Later, a polymorphism in the sequence of the splicing donor site of the third intron (G/A) was found among B. rapa accessions, and the presence of accessions encoding the complete BrFLC5 gene was reported (Xi et al. 2018). Quantitative trait loci (QTLs) affecting flowering time in different populations of B. rapa have been identified (Shea et al. 2018; Akter et al. 2021), and some QTLs cover regions including BrFLC1, BrFLC2, or VERNALIZATION INSENSITIVE3.1 (BrVIN3.1) (Zhao et al. 2010; Su et al. 2018). A flowering time QTL covering BrFLC5 was identified (Kakizaki et al. 2011), and the possibility that BrFLC5 is involved in flowering time variation has been shown (Kakizaki et al. 2011; Xi et al. 2018).
An accession encoding a complete BrFLC5 has been identified, and there is a flowering time difference between plants having a functional (G) and non-functional (A) polymorphism at the 5′ splice site in the third intron of BrFLC5 in the F2 population, suggesting that BrFLC5 contributes to the flowering time difference (Xi et al. 2018). Here, we identified accessions having a functional (G) or non-functional (A) polymorphism at the 5′ splice site in the third intron of BrFLC5 from over 300 B. rapa accessions. We confirmed that BrFLC5 acts as a floral repressor by overexpressing BrFLC5 in Arabidopsis thaliana. The variation of expression levels of BrFLC5 in pre-vernalized plants among B. rapa accessions and the variation in the repression rate of BrFLC5 expression following prolonged cold treatment were examined. Finally, we discuss whether BrFLC5 can be used as a breeding resource.
Materials and methods
Plant materials and growth conditions
Three hundred ten B. rapa accessions from four different varieties were used to examine a polymorphism in the sequence of the splicing donor site of the third intron (G/A) (Kawamura et al. 2016) (Table S1). Ten B. rapa accessions (Chinese cabbage; Chukanbohon Nou 6 gou (Nou 6) (Kakizaki et al. 2011), “Eishun,” “Nozaki Hakusai No. 2” (Nozaki Saishujo Ltd., Japan), “CR Kisaku-80,” “Raiou-90” (Marutane Co., Ltd., Japan), “CR-Okiniiri,” “Homarenokiwami,” “Shoshun” (Takii & Co., Ltd., Japan), Turnip; “Aishinku No. 3,” “CR Takamaru” (Musashino Seed Co., Ltd., Japan)) were used to examine the expression of BrFLC5 in pre-vernalized condition. After surface sterilization, the seeds were sown in agar-solidified Murashige and Skoog (MS) plates with 1% (w/v) sucrose under long-day (LD) conditions (16-h light) at 22 °C. Fourteen-day seedlings on MS plate were treated for 4 weeks at 4 °C under LD conditions. F2 seeds were produced by self-pollinating by bud pollination of F1 crossed by Nou 6 and “Eishun.” F2 seeds were sown in Petri dishes containing moistened paper and then transplanted into cell trays filled with soil. At 14 days after sowing, one leaf was used for RNA extraction and the remaining leaves for DNA extraction.
Sequencing DNA fragments of BrFLC5 gene
Genomic DNAs were isolated from 14-day first and second leaves by the cetyl trimethyl ammonium bromide (CTAB) method (Murray and Thompson 1980). The regions including the third intron were amplified by PCR using a primer set, BrFLC5-F/R (Fig. S1; Table S2). Quick Taq® HS DyeMix (Toyobo Co., Ltd., Osaka, Japan) was used for PCR. Amplified PCR products were treated by illustra ExoProStar (GE Healthcare Life Sciences, Chicago, Illinois, USA) and were directly sequenced using ABI Prism 3130 (Applied Biosystems, Foster City, California, USA).
The 1355- or 771-bp upstream regions from the transcription start site (TSS) were amplified by PCR using a primer set, BrFLC5_pro-F1/R1 (Fig. S1; Table S2). Amplified PCR fragments (771 bp) of seven accessions (“Nozaki Hakusai No. 2,” “CR Kisaku-80,” “Raiou-90,” “Eishun,” “Shoshun”, “Homarenokiwami,” and “CR-Okiniiri”) were directly sequenced. Amplified PCR products (1355 bp) of three accessions (Nou 6, “Aishinku No. 3,” and “CR Takamaru”) were cloned into pGEM®-T Easy vector (Promega, Madison, WI, USA). Quick Taq® HS DyeMix (Toyobo) was used for PCR. Nucleotide sequences of five clones of PCR products were determined with the ABI Prism 3130 (Applied Biosystems). Primers, M13F/R and BrFLC5_pro F2/R2, were used for sequencing (Fig. S1; Table S2). The data were analyzed using Sequencher (Gene Codes Corporation, MI, USA). Genotyping of F2 plants derived from Nou 6 and “Eishun” was performed by PCR using a primer set, BrFLC5_pro-F1/R1 (Fig. S1; Table S2). PCR was performed using the following conditions: 1 cycle of 94 °C for 2 min, 35 cycles of 94 °C for 30 s, 58 °C for 30 s, and 68 °C for 2 min. Primer sequences and their position used for sequencing are shown in Fig. S1 and Table S2.
RNA extraction and RT-qPCR
Total RNA was isolated from 14-day first and second leaves or from first and second leaves following 4 weeks of cold treatments to 14-day seedling using the SV Total RNA Isolation System (Promega). The leaves from three individual plants in each condition were harvested as biological replicates. The cDNA was synthesized from 500 ng total RNA using ReverTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo). RT-qPCR was performed using LightCycler 96 (Roche Diagnostics International Ltd., Switzerland). The cDNA was amplified using FastStart Essential DNA Green Master (Roche). PCR conditions were 95 °C for 10 min followed by 40 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 15 s, and the melting program (60 °C to 95 °C at 0.1 °C/s). After amplification cycles, each reaction was subjected to melt temperature analysis to confirm single amplified products. The expression level of each gene relative to BrACTIN was automatically calculated using automatic CQ calling according to the manufacturer’s instructions (Roche) (Fujimoto et al. 2006). Data presented are the average and standard error (s.e.) calculated from three biological and experimental replications. Primer sequences used for RT-qPCR are shown in Table S2.
RNA-sequencing
For RNA-sequencing (RNA-seq), total RNA from 14-day first and second leaves of pre-vernalized in “Aishinku No. 3,” “CR Takamaru,” Nou 6, and “Homarenokiwami” were used. A library was prepared using NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, MA, USA), and sequencing was performed using NovaSeq 6000 (Illumina Inc., San Diego, CA) (paired-end, 150 bp). Low-quality reads were filtered using FaQCs ver 2.10 (Lo and Chain 2014), and HISAT2 ver 2.2.1 (Kim et al. 2015) was used to align the filtered reads to Brassica rapa reference sequence v1.5 (http://brassicadb.org/cn/). The levels of gene expression were scored by fragments per kilo-base per million (FPKM) using cuffdiff v2.2.1 (Trapnell et al. 2010). The RNA-seq data were deposited in the DNA Data Bank of Japan (DDBJ; accession no. DRA015525)
Constructs and plant transformation
The cDNA from leaves of Nou 6 was used for RT-PCR. PrimeSTAR GXL DNA Polymerase (Takara Bio, Shiga, Japan) was used for RT-PCR. PCR fragments of a coding sequence (CDS) of BrFLC5 were amplified by RT-PCR using a primer pair, BrFLC5_5UTR_Sal and BrFLC5_3UTR_XbaI-2 (Table S2). PCR was performed using the following conditions: 1 cycle of 94 °C for 2 min, 35 cycles of 94 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s. Amplified PCR products were then cloned into the pGEM®-T Easy vector (Promega). The Sal I and Xba I fragment including BrFLC5 CDS was inserted between the 35S promoter and Nos terminator in the overexpression vector prepared by modifying the pCAMBIA1301 binary vector (Itabashi et al. 2019). Ligation was carried out using In-Fusion HD Cloning Kit (Takara Bio), and a primer pair, BrFLC5_CDS-fusF3 and BrFLC5_CDS-fusR3, was used (Table S2). This construct was transformed into Agrobacterium tumefaciens strain EHA105, and the transformation of Columbia-0 (Col) accession in A. thaliana was carried out by the floral dip procedure (Clough and Bent 1998). Transgenic seedlings were selected, which showed hygromycin resistance on a selection medium. T2 plant seeds were sown on MS medium and grown under LD conditions (16-h light) at 22 °C. After growing plants on MS medium, they were transferred to soil and grown under the conditions described above. The flowering time observed in A. thaliana was expressed as the number of rosette leaves at the time of flowering. Total RNA was isolated from mature leaves of T2 plants with and without transgene, and RT-PCR was performed to confirm the expression of the transformed BrFLC5 gene. PCR conditions were 1 cycle of 94 °C for 2 min, 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 68 °C for 30 s. Primer sequences used for RT-PCR are shown in Table S2.
Results
Identification of functional and non-functional alleles in B. rapa germplasm
In the reference genome sequence of B. rapa, Chiifu-401-42, BrFLC5 (Bra022771/BraA03g015950.3C) is annotated with five exons, while the other three BrFLC paralogs (BrFLC1, BrFLC2, and BrFLC3) are annotated with seven exons (Wang et al. 2011; Zhang et al. 2022), implying that BrFLC5 is a pseudogene in B. rapa. The reason for the decrease in the number of exons is the presence of a mutation from G to A in the splicing donor site (GT) in the third intron (Fig. S1). However, some lines of B. rapa have G nucleotide in the splicing donor site; in these accessions, there are seven exons in BrFLC5 (Xi et al. 2018). In this study, we examined the G/A polymorphism in 310 accessions of B. rapa that were mainly collected from Japanese commercial cultivars. Direct sequencing of the 673 bp sequence harboring this G/A polymorphism was performed in 310 B. rapa accessions (Fig. S1). Among the 310 accessions, 19 accessions (6.1%) had a homozygous G/G allele, 81 accessions (26.1%) had a heterozygous G/A allele, and 210 accessions (67.7%) had a homozygous A/A allele (Table 1). Among four varieties of B. rapa, the frequency of homozygous G/G alleles was 31.3% in turnip, 5.0% in komatsuna, 4.8% in Chinese cabbage, and none in pak choi. In all varieties, the frequency of a homozygous G/G allele was lower than that of a homozygous A/A allele (Table 1).
The reason for the different frequency of a functional BrFLC5 allele between Xi et al. (2018) and our study is that our plant materials included a higher percentage of Chinese cabbage accessions, and Xi et al. (2018) included more turnip and pak choi accessions. The frequency of homozygous G/G alleles differed between the two plant material sets, and the plant materials of Xi et al. (2018) contained a higher frequency of homozygous G/G alleles than ours. Turnip accessions had the highest percentage of homozygous G/G alleles in both plant materials (Fig. S2).
BrFLC5 functions as a floral repressor
BrFLC1, BrFLC2, and BrFLC3 but not BrFLC5 have been shown to act as floral repressors (Kim et al. 2007; Takada et al. 2019). The nucleotide sequence of the coding region of BrFLC5 in Nou 6 was determined. The similarities of the predicted amino acid sequence of BrFLC5 to BrFLC1, BrFLC2, and BrFLC3 were 81%, 84%, and 80%, respectively (Fig. 1). The MADS-box domain responsible for binding to target DNA was conserved in BrFLC5 (Fig. 1).
A 35S promoter::BrFLC5 CDS was transformed into the Columbia-0 accession of A. thaliana that has no AtFLC expression because of the loss of function of FRIGIDA (AtFRI) (Johanson et al. 2000). More than twenty T1 plants were obtained. Some T1 plants did not flower and offspring were not obtained. Three T2 populations (T2-1, 2, and 3 populations) were examined for flowering time, and the flowering time of plants transgenic for BrFLC5 transgene (TG+) was later than plants without the transgene (TG−) (Fig. 2a, b). All plants transformed with transgene did not show delayed flowering. This was also observed in previous reports (Kim et al. 2007; Itabashi et al. 2019; Takada et al. 2019), and Kim et al. (2007) suggested that the protein level of FLC might be the cause. The flowering time of plants transgenic for BrFLC5 transgene (TG+) was also later than plants without the transgene (TG−) in two T3 populations; T3-1 and T3-2 populations were derived from T2-2 and T2-3 late flowering plants, respectively (Fig. S3). The expression of BrFLC5 in transgenic plants was confirmed by RT-PCR (Fig. 2c).
Overexpression of BrFLC5 causes late flowering. a Number of rosette leaves in three T2 populations that are overexpressing BrFLC5. b Flowering time phenotypes in T2 plants with overexpressing BrFLC5. No cold (4 °C) treatments were applied, and the plants were grown under long-day conditions (16-h light) at 22 °C. c RT-PCR analysis showing transcription of BrFLC5. Three independent plants from the T2-2 population were used. AtGAPD was used as a control of demonstrate equal concentration of cDNA templates. TG+ and TG− represent the presence and absence of transgenes (TG), respectively
Expression levels of BrFLC5 varied among B. rapa accessions
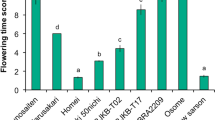
We have suggested that the level of FLC expression prior to prolonged cold treatment and the rate of suppression of FLC expression following prolonged cold treatment are important in the vernalization requirement in B. rapa (Takada et al. 2019; Akter et al. 2021). The expression level of BrFLC5 was examined by qPCR using a BrFLC5-specific primer set in ten accessions of B. rapa of which seven had homozygous G/G alleles and three had homozygous A/A alleles. “Aishinku No. 3” showed the highest expression levels followed by “CR Takamaru” and Nou 6, which all have a homozygous G/G allele (Fig. 3). The expression levels of “Homarenokiwami” and “CR-Okiniiri,” which have the homozygous A/A allele, were similar to that in “Raiou-90” and higher than those in “Nozaki Hakusai No. 2,” “CR Kisaku-80,” and “Eishun” that all have a homozygous G/G allele (Fig. 3), indicating that the expression level of BrFLC5 in pre-vernalized plants was independent of the G/A polymorphism in its third intron.
The expression levels of BrFLC5 with (4V) and without (NV) 4 weeks of cold treatments. The expression level of BrFLC5 relative to BrACTIN was calculated. Data presented are average and standard error (s.e.) of three biological and experimental replications. Percentages (%) shown above the bars are the ratio of expression level in 4V samples compared with NV samples. NV, pre-vernalized; 4V, 4 weeks of vernalized; G, functional allele; A, non-functional allele. The letters above the bars represent significant differences at p<0.05 (Tukey-Kramer test). Type A has no insertion in the promoter region and has an A/A homozygous allele. Type B has no insertion in the promoter region and has a G/G homozygous allele. Type C has a 584-bp insertion in the promoter region and has a G/G homozygous allele. 1, “Nozaki Hakusai No. 2”; 2, “CR Kisaku-80”; 3, “Eishun”; 4, “Raiou-90”; 5, Nou 6; 6, “Aishinku No. 3”; 7, “CR Takamaru”; 8, “Shoshun”; 9, “Homarenokiwami”; 10, “CR-Okiniiri”
The BrFLC5 promoter sequences of ten accessions of B. rapa were determined. Among these ten accessions, there were two fragment sizes of promoter regions, 1335 bp (type C, G/G allele) and 771 bp (type A, A/A allele; type B, G/G allele) (Figs. 3 and 4). There were no sequence differences among accessions having the same fragment sizes, and the 771-bp sequence was identical to the sequences in the reference genome. The 1335-bp fragment has 584-bp insertion (Fig. 4), and all accessions having this insertion tended to have higher expression levels of BrFLC5 before vernalization (Figs. 3 and 4). This 584-bp inserted region has an AT-rich sequence in its central region, and tandem duplications were found in this region (Fig. S4). Blast searches using this 584-bp sequence as a query revealed a homologous sequence in the promoter region of FLC.A03b of B. napus (Identity 583/584, 99.8%) that is an ortholog of BrFLC5 (Akter et al. 2021); there were many regions homologous to this 584-bp sequence in the genome of B. rapa (Fig. S5).
Two types of fragment sizes in the promoter region of BrFLC5. Type A has no insertion in the promoter region and has an A/A homozygous allele and includes three accessions (“Shoshun,” “Homarenokiwami,” “CR-Okiniiri”). Type B has no insertion in the promoter region and has a G/G homozygous allele including four accessions (“Nozaki Hakusai No. 2,” “CR Kisaku-80,” “Eishun,” “Raiou-90”). Type C has 584-bp insertion in the promoter region and a higher expression level of BrFLC5 including in three accessions (Nou 6, “Aishinku No. 3,” “CR Takamaru”). Gray box represents the insertion and black boxes indicate the exons. G, putative functional allele; A, non-functional allele
An F2 population was produced by crossing Nou 6 (type C) and “Eishun” (type B), and eight F2 plants homozygous for the 584-bp insertion or without the 584-bp insertion were selected. The expression levels of BrFLC5 of the eight F2 plants with a homozygous 584-bp insertion were higher than those without the 584-bp insertion (Fig. S6), suggesting that this 584-bp insertion is associated with a higher BrFLC5 expression level.
In response to 4 weeks of cold treatment, BrFLC5 expression levels decreased to between 6.2 and 39.4% of the non-vernalized level, and the average repression rate in accessions having G/G homozygous (25.1%) was similar to those having A/A homozygous (24.3%) (Fig. 3).
BrFLC5showed the lowest expression level among four BrFLC paralogs
We performed RNA-seq in the 14-day first and second leaves in four accessions of B. rapa (Nou 6, “Aishinku No. 3,” “CR Takamaru,” and “Homarenokiwami”); three of the four accessions (Nou 6, “Aishinku No. 3,” and “CR Takamaru”) have a homozygous G/G allele with 584-bp insertion in the BrFLC5 promoter region. From 82.2 to 92.1% of total filtered reads (15.5- to 21.2-M reads) were mapped to the reference genome (Table S3). Expression levels of BrFLC1, BrFLC2, BrFLC3, and BrFLC5 were identified using the FPKM value. BrFLC3 had the highest expression levels in all four accessions, from 40.1 to 56.9% of the total BrFLC expression level, and BrFLC5 had the lowest expression levels, from 2.4 to 6.2% of the total BrFLC expression level (Fig. 5).
Discussion
Flowering time is an important trait for B. rapa vegetables, and FLC plays an important role in controlling flowering time in B. rapa (Akter et al. 2021). BrFLC5 originated from the α replication of the common ancestor of A. thaliana and B. rapa, but it was not localized in the syntenic region where AtFLC is located (Yang et al. 2006; Akter et al. 2019). In the reference genome of B. rapa, BrFLC5 was considered to be a pseudogene because it lacks two exons (Schranz et al. 2002; Wang et al. 2011). Furthermore, the expression level of BrFLC5 is the lowest among the four paralogs in one accession (Xi et al. 2018), and in this study, similar results were found in four accessions. BoFLC5 can also be a pseudogene and has the lowest expression levels among the paralogs in cabbage (Brassica oleracea) (Itabashi et al. 2019). The expression levels of BnaFLC.A03b and BnaFLC.C03b, which are orthologs of BrFLC5 and BoFLC5, respectively, tended to be lower than the other BnFLC paralogs in B. napus (Calderwood et al. 2021). These results suggest that sequences that are associated with low expression levels of FLC5 were fixed early after the speciation of Brassica species.
The contribution of functional BrFLC5 to flowering time variation was shown using the F2 population derived from accessions having A homozygous and G homozygous alleles (Xi et al. 2018); BrFLC5 functions weakly to control flowering time even though other BrFLC paralogs affect the flowering time. QTL analysis using Nou 6 and A9709, which are high and low vernalization requirements, respectively, was performed in field conditions, sowing seeds in the winter season in Japan (January or February) without any cold treatment. Two QTLs were identified and one of them covered BrFLC5 (Kakizaki et al. 2011). In this study, we confirmed that A9709 has a homozygous A allele, type A, and Nou 6 has a homozygous G allele, type C. Based on the data of Kakizaki et al. 2011 in four environmental conditions, the F2 plants having type C BrFLC5 homozygous allele tended to have shorter stem length (higher vernalization requirement/later flowering time) than F2 plants having type A BrFLC5 homozygous allele when BrFLC1 allele was fixed (Fig. S7), suggesting that some of the flowering time difference of these two accessions might be due to the BrFLC5 function; differences between type A and type C of the BrFLC5 allele were shown to be possibly related to differences in vernalization requirement (flowering time) (Kakizaki et al. 2011). In this study, we confirmed that BrFLC5 acts as a floral repressor. Integrating these results suggests that the functional difference of BrFLC5 (A allele, type A vs. G allele, type B/C) is associated with the difference in vernalization requirement/bolting time in B. rapa.
There are a few studies examining the relationship between BrFLC5 and flowering time/vernalization requirement. However, some accessions having a full BrFLC5 amino acid sequence have been identified (Xi et al. 2018). We also identified some accessions with full annotation of the BrFLC5 amino acid sequence among 310 B. rapa accessions. Despite the differences in the populations used, the two studies showed that the frequency of the A allele (non-functional) is higher than the G allele (putative functional) and turnip showed the highest ratio of homozygous G allele. These results may be due to the stronger selection for vernalization requirements in turnip than in other B. rapa vegetables.
Transposable elements (TEs) have been found in the genomic regions in FLC in B. rapa and B. napus and sometimes affect the FLC expression levels before vernalization or the repression rate of FLC expression following prolonged cold treatments (Akter et al. 2021). We found a 584-bp insertion in the promoter region, and this sequence did not show any structures that would be considered TE but had tandem duplications. Accessions having this insertion tended to have higher BrFLC5 expression levels than accessions without this insertion in pre-vernalized plants, though this insertion did not affect the repression of BrFLC5 expression following prolonged cold treatments. High BrFLC5 expression levels were found to be linked to this insertion using an F2 population derived from crossed accessions having homozygous type B and type C BrFLC5 alleles. Though the possibility that the other sequence variations of BrFLC5 between parental lines of the F2 population results in the difference of BrFLC5 expression was not completely excluded, we suggest that this insertion leads to increased BrFLC5 expression in pre-vernalized plants. However, it is necessary to test for association using more accessions, to test for association using segregating populations, or to prove it by promoter assay.
Leafy and root B. rapa vegetables need a high requirement for vernalization to prevent premature bolting, which reduces the yield and quality of products (Akter et al. 2021). A higher expression level before vernalization is one important characteristic of a high vernalization requirement (Takada et al. 2019; Akter et al. 2021); thus, the higher functional BrFLC5 expression level in pre-vernalized material like type C BrFLC5 allele could be important for a high vernalization requirement. A variation of BrFLC5 expression levels among accessions having functional BrFLC5 alleles, type B and type C, in pre-vernalized plants was observed in this study. Thus, if there is an association between this insertion and increased BrFLC5 expression before vernalization, developing a DNA marker identifying this insertion will be useful for marker-assisted selection.
Conclusion
Some accessions have a functional BrFLC5 allele, and this functional difference between varieties may be associated with differences in vernalization requirement in B. rapa. There is a variation of BrFLC5 expression levels between accessions prior to vernalization. Though the effect may not be significant due to BrFLC5 having the lowest expression levels among the four FLC paralogs, higher functional BrFLC5 expression might contribute to increased vernalization requirement. Thus, the type C allele with a functional BrFLC5 and higher BrFLC5 expression levels may be useful for developing high bolting-resistant breeding in B. rapa vegetables.
Data availability
DDBJ; accession no. DRA015525.
References
Akter A, Itabashi E, Kakizaki T, Okazaki K, Dennis ES, Fujimoto R (2021) Genome triplication leads to transcriptional divergence of FLOWERING LOCUS C genes during vernalization in the genus Brassica. Front Plant Sci 11:619417. https://doi.org/10.3389/fpls.2020.619417
Akter A, Miyazaki J, Shea DJ et al (2020) Gene expression analysis in response to vernalization in Chinese cabbage (Brassica rapa L.). Hort J 89:268–277. https://doi.org/10.2503/hortj.UTD-150
Akter A, Nishida N, Takada S, Itabashi E, Osabe K, Shea DJ, Fujimoto R (2018) Genetic and epigenetic regulation of vernalization in Brassicaceae. In: El-Esawi MA (ed) Brassica germplasm–characterization, breeding and utilization. IntechOpen, London, pp 75–94
Akter A, Takahashi S, Deng W et al (2019) The histone modification H3 lysine 27 tri-methylation has conserved gene regulatory roles in the triplicated genome of Brassica rapa L. DNA Res 26:433–443. https://doi.org/10.1093/dnares/dsz021
Belser C, Istace B, Denis E et al (2018) Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat Plants 4:879–887. https://doi.org/10.1038/s41477-018-0289-4
Calderwood A, Lloyd A, Hepworth J et al (2021) Total FLC transcript dynamics from divergent paralogue expression explains flowering diversity in Brassica napus. New Phytol 229:3534–3548. https://doi.org/10.1111/nph.17131
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Fujimoto R, Sasaki T, Nishio T (2006) Characterization of DNA methyltransferase genes in Brassica rapa. Genes Genet Syst 81:235–242. https://doi.org/10.1266/ggs.81.235
Itabashi E, Osabe K, Fujimoto R, Kakizaki T (2018) Epigenetic regulation of agronomical traits in Brassicaceae. Plant Cell Rep 37:87–101. https://doi.org/10.1007/s00299-017-2223-z
Itabashi E, Shea DJ, Fukino N, Fujimoto R, Okazaki K, Kakizaki T, Ohara T (2019) Comparison of cold responses for orthologs of cabbage vernalization-related genes. Hort J 88:462–470. https://doi.org/10.2503/hortj.UTD-059
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347. https://doi.org/10.1126/science.290.5490.344
Kakizaki T, Kato T, Fukino N, Ishida M, Hatakeyama K, Matsumoto S (2011) Identification of quantitative trait loci controlling late bolting in Chinese cabbage (Brassica rapa L.) parental line Nou 6 gou. Breed Sci 61:151–159. https://doi.org/10.1270/jsbbs.61.151
Kawamura K, Kawanabe T, Shimizu M et al (2016) Genetic distance of inbred lines of Chinese cabbage and its relationship to heterosis. Plant Gene 5:1–7. https://doi.org/10.1016/j.plgene.2015.10.003
Kawanabe T, Osabe K, Itabashi E, Okazaki K, Dennis ES, Fujimoto R (2016) Development of primer sets that can verify the enrichment of histone modifications, and their application to examining vernalization-mediated chromatin changes in Brassica rapa L. Genes Genet Syst 91:1–10. https://doi.org/10.1266/ggs.15-00058
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Kim SY, Park BS, Kwon SJ et al (2007) Delayed flowering time in Arabidopsis and Brassica rapa by the overexpression of FLOWERING LOCUS C (FLC) homologs isolated from Chinese cabbage (Brassica rapa L. ssp. pekinensis). Plant Cell Rep 26:327–336. https://doi.org/10.1007/s00299-006-0243-1
Kitamoto N, Yui S, Nishikawa K, Takahata Y, Yokoi S (2014) A naturally occurring long insertion in the first intron in the Brassica rapa FLC2 gene causes delayed bolting. Euphytica 196:213–223. https://doi.org/10.1007/s10681-013-1025-9
Li F, Kitashiba H, Inaba K, Nishio T (2009) A Brassica rapa linkage map of EST-based SNP markers for identification of candidate genes controlling flowering time and leaf morphological traits. DNA Res 16:311–323. https://doi.org/10.1093/dnares/dsp020
Li P, Su T, Zhao X et al (2021) Assembly of the non-heading pak choi genome and comparison with the genomes of heading Chinese cabbage and the oilseed yellow sarson. Plant Biotechnol J 19:966–976. https://doi.org/10.1111/pbi.13522
Li Y, Liu GF, Ma LM et al (2020) A chromosome-level reference genome of non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hortic Res 7:212. https://doi.org/10.1038/s41438-020-00449-z
Lo CC, Chain PSG (2014) Rapid evaluation and quality control of next generation sequencing data with FaQCs. BMC Bioinform 15:366. https://doi.org/10.1186/s12859-014-0366-2
Lv H, Miyaji N, Osabe K, Akter A, Mehraj H, Shea DJ, Fujimoto R (2020) The importance of genetic and epigenetic research in the Brassica vegetables in the face of climate change. In: Kole C (ed) Genomic designing of climate-smart vegetable crops. Springer, Cham, pp 161–255
Mehraj H, Takahashi S, Miyaji N et al (2021) Characterization of histone H3 lysine 4 and 36 tri-methylation in Brassica rapa L. Front Plant Sci 12:659634. https://doi.org/10.3389/fpls.2021.659634
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326. https://doi.org/10.1093/nar/8.19.4321
Qi X, An H, Ragsdale AP, Hall TE, Gutenkunst RN, Pires JC, Barker MS (2017) Genomic inferences of domestication events are corroborated by written records in Brassica rapa. Mol Ecol 26:3373–3388. https://doi.org/10.1111/mec.14131
Schranz ME, Quijada P, Sung SB, Lukens L, Amasino R, Osborn TC (2002) Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162:1457–1468. https://doi.org/10.1093/genetics/162.3.1457
Shea DJ, Itabashi E, Takada S, Fukai E, Kakizaki T, Fujimoto R, Okazaki K (2018) The role of FLOWERING LOCUS C in vernalization of Brassica: the importance of vernalization research in the face of climate change. Crop Pasture Sci 69:30–39. https://doi.org/10.1071/CP16468
Shea DJ, Nishida N, Takada S et al (2019) Long noncoding RNAs in Brassica rapa L. following vernalization. Sci Rep 9:9302. https://doi.org/10.1038/s41598-019-45650-w
Su T, Wang W, Li P et al (2018) A genomic variation map provides insights into the genetic basis of spring Chinese cabbage (Brassica rapa ssp. pekinensis) selection. Mol Plant 11:1360–1376. https://doi.org/10.1016/j.molp.2018.08.006
Takada S, Akter A, Itabashi E et al (2019) The role of FRIGIDA and FLOWERING LOCUS C genes in flowering time of Brassica rapa leafy vegetables. Sci Rep 9:13843. https://doi.org/10.1038/s41598-019-50122-2
Trapnell C, Williams BA, Pertea G et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515. https://doi.org/10.1038/nbt.1621
Wang X, Wang H, Wang J et al (2011) The genome of the mesopolyploid crop species Brassica rapa. Nat Genet 43:1035–1039. https://doi.org/10.1038/ng.919
Xi X, Wei K, Gao B et al (2018) BrFLC5: a weak regulator of flowering time in Brassica rapa. Theor Appl Genet 131:2107–2116. https://doi.org/10.1007/s00122-018-3139-x
Yang TJ, Kim JS, Kwon SJ et al (2006) Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18:1339–1347. https://doi.org/10.1105/tpc.105.040535
Yuan YX, Wu J, Sun RF, Zhang XW, Xu DH, Bonnema G, Wang XW (2009) A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J Exp Bot 60:1299–1308. https://doi.org/10.1093/jxb/erp010
Zhang L, Cai X, Wu J et al (2018) Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res 5:50. https://doi.org/10.1038/s41438-018-0071-9
Zhang Z, Guo J, Cai X et al (2022) Improved reference genome annotation of Brassica rapa by Pacific Biosciences RNA sequencing. Front Plant Sci 13:841618. https://doi.org/10.3389/fpls.2022.841618
Zhao J, Kulkarni V, Liu N, Del Carpio DP, Bucher J, Bonnema G (2010) BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J Exp Bot 61:1817–1825. https://doi.org/10.1093/jxb/erq048
Acknowledgements
We thank Ms. Tomoko Kusumi and Ms. Yumiko Arai for their technical assistance.
Funding
Open access funding provided by Kobe University. This work was supported by the Kobe University Strategic International Collaborative Research Grant (Type B Fostering Joint Research), Fund for the Promotion of Joint Research (16KK0171), and Grant-in-Aid for Scientific Research (B) (18H02173, 21H02163) of Japan Society for the Promotion of Science (JSPS) granted to R.F.
Author information
Authors and Affiliations
Contributions
AA, TK, EI, KK, MAA, and HM performed experiments, and AA, EI, and MS performed data analysis. AA, ESD, and RF drafted the manuscript. AA, TK, KO, and RF participated in the design of the study. ESD and RF revised the manuscript. All of the authors have read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Akter, A., Kakizaki, T., Itabashi, E. et al. Characterization of FLOWERING LOCUS C 5 in Brassica rapa L.. Mol Breeding 43, 58 (2023). https://doi.org/10.1007/s11032-023-01405-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-023-01405-0