Abstract
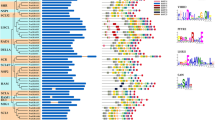
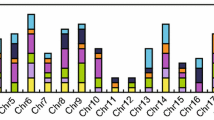
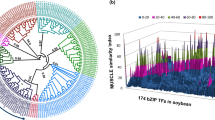
The BYPASS1-related gene (BPS1) encodes a protein with an unknown functional domain that regulates plant organ growth and development by inhibiting the continuous production of a root-derived long-distance signaling molecule called bypass (bps). We conducted a comprehensive study to investigate the BPS gene family in soybean and identified twenty-three BPS genes in Glycine max and twenty BPS genes in Glycine soja (wild soybean). Collinearity analysis revealied the existence of multiple orthologs of soybean BPS genes in wild soybean, indicating incomplete conservation between the BPS genes of soybean and wild soybean. Phylogenetic analysis successfully categorized all BPS genes into five distinct groups. We further scrutinized their chromosomal locations, gene structures, conserved motifs, cis-acting elements, and expression patterns. Leveraging publicly available data on genetic variation, phenotypic variation, and single-cell transcriptome sequencing of root nodules, we discovered a potential association between BPS genes and multiple soybean traits, particularly those related to the root nodule phenotype. This pioneering study provides a systematic and comprehensive examination of the BPS gene family in soybean. The findings establish a robust foundation for future investigations into the functional roles of BPS genes in plant growth and development.






Similar content being viewed by others
Data Availability
All data used in this paper that has been released.
References
Ablazov A, Mi J, Jamil M, Jia KP, Wang JY, Feng Q, Al-Babili S (2020) The apocarotenoid zaxinone is a positive regulator of strigolactone and abscisic acid biosynthesis in arabidopsis roots. Front Plant Sci 14(11):578
Adhikari E, Lee DK, Giavalisco P, Sieburth LE (2013) Long-distance signaling in bypass1 mutants: bioassay development reveals the bps signal to be a metabolite. Mol Plant 6(1):164–173
Arthikala MK, Nanjareddy K, Lara M (2018) In BPS1 Downregulated roots, the BYPASS1 signal disrupts the induction of cortical cell divisions in bean-Rhizobium symbiosis. Genes (Basel) 9(1):11
Bailey TL, Johnson J, Grant CE, Noble WS (2015) The MEME suite. Nucleic Acids Res 43(W1):W39–W49
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202
Choi WG, Miller G, Wallace I, Harper J, Mittler R, Gilroy S (2017) Orchestrating rapid long-distance signaling in plants with Ca2+. ROS and electrical signals Plant J 90(4):698–707
Christmann A, Grill E, Huang J (2013) Hydraulic signals in long-distance signaling. Curr Opin Plant Biol 16(3):293–300
Garg VK, Avashthi H, Tiwari A, Jain PA, Ramkete PW, Kayastha AM, Singh VK (2016) MFPPI - multi FASTA ProtParam interface. Bioinformation 12(2):74–77
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS (2012) Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res 40(Database issue):D1178–D1186
Hanada K, Zhang X, Borevitz JO, Li WH, Shiu SH (2007) A large number of novel coding small open reading frames in the intergenic regions of the Arabidopsis thaliana genome are transcribed and/or under purifying selection. Genome Res 17(5):632–640
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297
Kang YW, Kim RN, Cho HS, Kim WT, Choi D, Pai HS (2008) Silencing of a BYPASS1 homolog results in root-independent pleiotrophic developmental defects in Nicotiana benthamiana. Plant Mol Biol 68(4-5):423–437
Kondhare KR, Patil NS, Banerjee AK (2021) A historical overview of long-distance signalling in plants. J Exp Bot 72(12):4218–4236
Kudla J, Becker D, Grill E, Hedrich R, Hippler M, Kummer U, Parniske M, Romeis T, Schumacher K (2018) Advances and current challenges in calcium signaling. New Phytol 218(2):414–431
Kulich I, Cole R, Drdová E, Cvrcková F, Soukup A, Fowler J, Zárský V (2010) Arabidopsis exocyst subunits SEC8 and EXO70A1 and exocyst interactor ROH1 are involved in the localized deposition of seed coat pectin. New Phytol 188(2):615–625
Lee DK, Sieburth LE (2012) The bps signal: embryonic arrest from an auxin-independent mechanism in bypass triple mutants. Plant Signal Behav 7(6):698–700
Lee DK, Parrott DL, Adhikari E, Fraser N, Sieburth LE (2016) The mobile bypass signal arrests shoot growth by disrupting shoot apical meristem maintenance, cytokinin signaling, and WUS transcription factor expression. Plant Physiol 171(3):2178–2190
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Liu Y, Du H, Li P, Shen Y, Peng H, Liu S, Zhou GA, Zhang H, Liu Z, Shi M, Huang X, Li Y, Zhang M, Wang Z, Zhu B, Han B, Liang C, Tian Z (2020) Pan-genome of wild and cultivated soybeans. Cell 182(1):162–176.e13
Liu Y, Zhang Y, Liu X, Shen Y, Tian D, Yang X, Liu S, Ni L, Zhang Z, Song S, Tian Z (2023b) SoyOmics: a deeply integrated database on soybean multi-omics. Mol Plant S1674-2052(23):00075–00078
Liu Z, Kong X, Long Y, Liu S, Zhang H, Jia J, Cui W, Zhang Z, Song X, Qiu L, Zhai J, Yan Z (2023a) Integrated single-nucleus and spatial transcriptomics captures transitional states in soybean nodule maturation. Nat Plants 9(4):515–524
Luo H, He W, Li D, Bao Y, Riaz A, Xiao Y, Song J, Liu C (2020) Effect of methyl jasmonate on carotenoids biosynthesis in germinated maize kernels. Food Chem 1(307):125525
Moreau C, Gautrat P, Frugier F (2021) Nitrate-induced CLE35 signaling peptides inhibit nodulation through the SUNN receptor and miR2111 repression. Plant Physiol 185(3):1216–1228
Mouchel CF, Leyser O (2007) Novel phytohormones involved in long-range signaling. Curr Opin Plant Biol 10(5):473–476
Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (2014) The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front Plant Sci 16(5):170
Putterill J, Varkonyi-Gasic E (2016) FT and florigen long-distance flowering control in plants. Curr Opin Plant Biol 33:77–82
Rehman NU, Ali M, Ahmad MZ, Liang G, Zhao J (2018) Strigolactones promote rhizobia interaction and increase nodulation in soybean (Glycine max). Microb Pathog 114:420–430
Roy S, Liu W, Nandety RS, Crook A, Mysore KS, Pislariu CI, Frugoli J, Dickstein R, Udvardi MK (2020) Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32(1):15–41
Savojardo C, Martelli PL, Fariselli P, Profiti G, Casadio R (2018) BUSCA: an integrative web server to predict subcellular localization of proteins. Nucleic Acids Res 46(W1):W459–W466
Takahashi F, Shinozaki K (2019) Long-distance signaling in plant stress response. Curr Opin Plant Biol 47:106–111
Turnbull C (2011) Long-distance regulation of flowering time. J Exp Bot 62(13):4399–4413
Van Norman JM, Sieburth LE (2007) Dissecting the biosynthetic pathway for the bypass1 root-derived signal. Plant J 49(4):619–628
Van Norman JM, Frederick RL, Sieburth LE (2004) BYPASS1 negatively regulates a root-derived signal that controls plant architecture. Curr Biol 14(19):1739–1746
Varsani S, Basu S, Williams WP, Felton GW, Luthe DS, Louis J (2016) Intraplant communication in maize contributes to defense against insects. Plant Signal Behav 11(8):e1212800
Wang X, Chen K, Zhou M, Gao Y, Huang H, Liu C, Fan Y, Fan Z, Wang Y, Li X (2022) GmNAC181 promotes symbiotic nodulation and salt tolerance of nodulation by directly regulating GmNINa expression in soybean. New Phytol 236(2):656–670
Wheeldon CD, Bennett T (2021) There and back again: an evolutionary perspective on long-distance coordination of plant growth and development. Semin Cell Dev Biol 109:55–67
Yang Y, Guo Y (2018) Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol 217(2):523–539
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grants 32090064 to F.K.) and the National Natural Science Foundation of China (32072086).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Fanjiang Kong and Lin Zhao designed the research; Xinxin Pei completed the bioinformatics analysis, wrote and revised the manuscript; Fan Wang, Haiping Du, and Milan He modified the manuscript; Lanxin Li, Chuanjie Gou, Zheng Chen, and Yanan Wang downloaded the data; and all authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Yes.
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Soybean Functional Genomics.
Supplementary information

Fig. s1
Haplotype analysis of GmBPS genes. (PNG 1084 kb)

Fig. s2
UAMP visualization of GmBPS genes in single-cell gene maps of soybean nodules. (PNG 444 kb)

Fig. s3
Barplot visualization of GmBPS genes in single-cell gene maps of soybean nodules. (PNG 3014 kb)
Table s1
Results of Ka/Ks analysis between GmBPS. (XLSX 39 kb)
Table s2
Blast results of GmBPS14 and GmBPS21 in publicly available soybean genomes. (XLSX 19 kb)
Table s3
Details of cis-acting elements in the promoter regions in Glycine spp., Arabidopsis and P.vulgaris. (XLSX 463 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pei, X., Wang, F., Du, H. et al. Genome-wide identification and functional prediction of BYPASS1-related (BPS1) homologs in soybean. Mol Breeding 43, 59 (2023). https://doi.org/10.1007/s11032-023-01403-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-023-01403-2