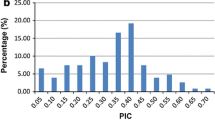
Abstract
Cotton is a major fiber and oilseed crop; however, few studies have been focused on oil content in cotton. To dissect the genetic basis of seed oil content (SOC), genome-wide association study was performed by using SOC phenotypic data of 503 upland cotton inbred accessions in eight environments and two sets of genotypic data including 179 simple sequence repeats (SSRs) and 11,975 single-nucleotide polymorphism (SNPs) from our previous study. SOC for a given genotype was fairly stable which was reflected in a broad-sense heritability of H2 = 96.6%. Using the mixed linear model (K + Q), 16 significant SSRs (P < 0.01) and 26 significant SNPs (P < 1/11,975), were detected cross BLUP values and at least one individual environments. Based on the LD block analysis, eight SOC-associated QTL on chromosome 6, 10, 12, 13, 15, 17, and 24 covering more than one significant markers were identified, two QTL of them on chromosome 10 and 13 could be matched between SSR and SNP markers. The confidence interval of candidate genes in each QTL ranged from 0.14 to 1.50 Mb, and 12 to 70 genes were identified in these QTL. According to the gene function annotation and gene FPKM value of the 20~35 days post anther (DPA) cotton ovules, key genes involved in lipid metabolism and transport; tricarboxylic acid cycle which might affect seed oil content were found near the leading markers. These results will facilitate the molecular marker assisted breeding of cotton oil content.





Similar content being viewed by others
References
Alfred Q, Liu HY, Xu HM, Li JR, Wu JG, Zhu SJ, Shi CH (2012) Mapping of quantitative trait loci for oil content in cottonseed kernel. J Genet 91:289–295
Bates PD, Stymne S, Ohlrogge J (2013) Biochemical pathways in seed oil synthesis. Curr Opin Plant Biol 16:358–364. https://doi.org/10.1016/j.pbi.2013.02.015
Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67:1–48. https://doi.org/10.18637/jss.v067.i01
Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, Napier JA, Kunst L (2009) Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme a reductase candidates of the fatty acid elongase. Plant Physiol 150:1174–1191. https://doi.org/10.1104/pp.109.137497
Dahlqvist A, Stahl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S (2000) Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-coa-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci U S A 97:6487–6492. https://doi.org/10.1073/pnas.120067297
de Silva K, Laska B, Brown C, Sederoff HW, Khodakovskaya M (2011) Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): a novel repressor of abiotic stress response. J Exp Bot 62:2679–2689. https://doi.org/10.1093/jxb/erq468
Dong XL, Bai PL, Wang JM, Ruan CJ (2011) Comparative study on determination of seed oil content of energy plants by using NMR and Soxhlet Extraction. Renew Energy Resour 29:21–24
Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M, Jia Y, Pan Z, Gong W, Liu Z, Zhu H, Ma L, Liu F, Yang D, Wang F, Fan W, Gong Q, Peng Z, Wang L, Wang X, Xu S, Shang H, Lu C, Zheng H, Huang S, Lin T, Zhu Y, Li F (2018a) Resequencing of 243 diploid cotton accessions based on an updated a genome identifies the genetic basis of key agronomic traits. Nat Genet 50(6):796–802. https://doi.org/10.1038/s41588-018-0116-x
Du X, Liu S, Sun J, Zhang G, Jia Y, Pan Z, Xiang H, He S, Xia Q, Xiao S, Shi W, Quan Z, Liu J, Ma J, Pang B, Wang L, Sun G, Gong W, Jenkins JN, Lou X, Zhu J, Xu H (2018b) Dissection of complicate genetic architecture and breeding perspective of cottonseed traits by genome-wide association study. BMC Genomics 19:451. https://doi.org/10.1186/s12864-018-4837-0
Elisabeth B, Urs S (2017) Evaluation of yield and drought using active and passive spectral sensing systems at the reproductive stage in wheat. Front Plant Sci 8:379. https://doi.org/10.3389/fpls.2017.00379
Fang M, Ding X, Li P, Zhang Z, Chen X, Jiang J, Zhang W, Gan D (2012) Modification of oilseeds soxhlet extraction for determination of oil content. Chin J Oil Crop Sci 34:210–214
Hamblin MT, Buckler ES, Jannink JL (2011) Population genetics of genomics-based crop improvement methods. Trends Genet 27:98–106. https://doi.org/10.1016/j.tig.2010.12.003
Holland JB, Nyquist WE, Cervantes-Martínez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112. https://doi.org/10.1002/9780470650202.ch2
Huang C, Nie X, Shen C, You C, Li W, Zhao W, Zhang X, Lin Z (2017) Population structure and genetic basis of the agronomic traits of upland cotton in China revealed by a genome-wide association study using high-density SNPs. Plant Biotechnol J 15:1374–1386. https://doi.org/10.1111/pbi.12722
Hulse-Kemp AM, Lemm J, Plieske J, Ashrafi H, Buyyarapu R, Fang DD, Frelichowski J, Giband M, Hague S, Hinze LL, Kochan KJ, Riggs PK, Scheffler JA, Udall JA, Ulloa M, Wang SS, Zhu QH, Bag SK, Bhardwaj A, Burke JJ, Byers RL, Claverie M, Gore MA, Harker DB, Islam MS, Jenkins JN, Jones DC, Lacape JM, Llewellyn DJ, Percy RG, Pepper AE, Poland JA, Mohan Rai K, Sawant SV, Singh SK, Spriggs A, Taylor JM, Wang F, Yourstone SM, Zheng X, Lawley CT, Ganal MW, Van Deynze A, Wilson IW, Stelly DM (2015) Development of a 63k SNP array for cotton and high-density mapping of intraspecific and interspecific populations of Gossypium spp. G3: Genes, Genomes, Genetics 5:1187–1209. https://doi.org/10.1534/g3.115.018416
Khan FA, Azhar FM, Afzal I, Irfanf S (2007) Effect of nitrogen regimes on combining ability variation in oil and protein contents in cottonseed (Gossypium hirsutum L.). Plant Prod Sci 10:367–371. https://doi.org/10.1626/pps.10.367
Liu HY, Quampah A, Chen JH, Li JR, Huang ZR, He QL, Shi CH, Zhu SJ (2012) QTL analysis for gossypol and protein contents in upland cottonseeds with two different genetic systems across environments. Euphytica 188:453–463. https://doi.org/10.1007/s10681-012-0733-x
Liu F, Zhao YP, Wang X, Li Y, Sun J (2015) Isolation and functional characterization of seed-specific fad2-1, promoter from cotton (Gossypium hirsutum L.). J Plant Biochem Biotechnol 24:369–375. https://doi.org/10.1007/s13562-014-0284-4
Liu H, Quampah A, Chen J, Li J, Huang Z, He Q, Shi C, Zhu S (2017) QTL mapping with different genetic systems for nine non-essential amino acids of cottonseeds. Mol Gen Genomics 292:671–684. https://doi.org/10.1007/s00438-017-1303-7
Mackay I, Powell W (2007) Methods for linkage disequilibrium mapping in crops. Trends Plant Sci 12:57–63. https://doi.org/10.1016/j.tplants.2006.12.001
Nie X, Huang C, You C, Li W, Zhao W, Shen C, Zhang B, Wang H, Yan Z, Dai B, Wang M, Zhang X, Lin Z (2016) Genome-wide SSR-based association mapping for fiber quality in nation-wide upland cotton inbreed cultivars in China. BMC Genomics 17:352. https://doi.org/10.1186/s12864-016-2662-x
Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL (2002) The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem 277:8877–8881. https://doi.org/10.1074/jbc.M111646200
Ramsay L, Comadran J, Druka A, Marshall DF, Thomas WT, Macaulay M, MacKenzie K, Simpson C, Fuller J, Bonar N, Hayes PM, Lundqvist U, Franckowiak JD, Close TJ, Muehlbauer GJ, Waugh R (2011) INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat Genet 43:169–172. https://doi.org/10.1038/ng.745
Shang L, Abduweli A, Wang Y, Hua J (2016) Genetic analysis and QTL mapping of oil content and seed index using two recombinant inbred lines and two backcross populations in upland cotton. Plant Breed 135:224–231. https://doi.org/10.1111/pbr.12352
Shockey JM, Fulda MS, Browse JA (2002) Arabidopsis contains nine long-chain acyl-coenzyme a synthetase genes that participate in fatty acid and glycerolipid metabolism. Plant Physiol 129:1710–1722. https://doi.org/10.1104/pp.003269
Sun S (1987) Study on the nutritional quality of cotton seeds. Sci Agric Sin 20:12–16
Sun Z, Wang X, Liu Z, Gu Q, Zhang Y, Li Z, Ke H, Yang J, Wu J, Wu L, Zhang G, Zhang C, Ma Z (2017) Genome-wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol J 15:982–996. https://doi.org/10.1111/pbi.12693
Wang Z, Yang C (2000) Determination of seed oil content of energy plants by using NMR methods. In: Chinese western cereals & oils technology, vol 25, pp 55–57
Ye Z, Lu Z, Zhu J (2003) Genetic analysis for developmental behavior of some seed quality traits in Upland cotton (Gossypum hirsutum L.). Euphytica 129:183–191. https://doi.org/10.1023/A:1021974901501
Yu J, Yu S, Fan S, Song M, Zhai H, Li X, Zhang J (2012) Mapping quantitative trait loci for cottonseed oil, protein and gossypol content in a Gossypium hirsutum × Gossypium barbadense, backcross inbred line population. Euphytica 187:191–201. https://doi.org/10.1007/s10681-012-0630-3
Zhang M, Fan J, Taylor DC, Ohlrogge JB (2009) DGAT1 and PDAT1 acyltransferases have overlapping functions in Arabidopsis triacylglycerol biosynthesis and are essential for normal pollen and seed development. Plant Cell 21:3885–3901. https://doi.org/10.1105/tpc.109.071795
Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, Hulse-Kemp AM, Wan Q, Liu B, Liu C, Wang S, Pan M, Wang Y, Wang D, Ye W, Chang L, Zhang W, Song Q, Kirkbride RC, Chen X, Dennis E, Llewellyn DJ, Peterson DG, Thaxton P, Jones DC, Wang Q, Xu X, Zhang H, Wu H, Zhou L, Mei G, Chen S, Tian Y, Xiang D, Li X, Ding J, Zuo Q, Tao L, Liu Y, Li J, Lin Y, Hui Y, Cao Z, Cai C, Zhu X, Jiang Z, Zhou B, Guo W, Li R, Chen ZJ (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33:531–537. https://doi.org/10.1038/nbt.3207
Zhao Y, Wang Y, Huang Y, Cui Y, Hua J (2018) Gene network of oil accumulation reveals expression profiles in developing embryos and fatty acid composition in upland cotton. J Plant Physiol 19:101–112. https://doi.org/10.1016/j.jplph.2018.06.002
Zheng H, Rowland O, Kunst L (2005) Disruptions of the Arabidopsis enoyl-coa reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17:1467–1481. https://doi.org/10.1105/tpc.104.030155
Zhu T, Liang C, Meng Z, Sun G, Meng Z, Guo S, Zhang R (2017) CottonFGD: an integrated functional genomics database for cotton. BMC Plant Biol 17:101. https://doi.org/10.1186/s12870-017-1039-x
Author contribution statement
ZL designed and conceived research. WZ, XK, and XN investigated phenotypic traits. WZ and YY conducted data analysis and wrote the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by National Key R&D Program of China (2018YFD0100405).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The authors declare that the experiments comply with the current laws of China.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, W., Kong, X., Yang, Y. et al. Association mapping seed kernel oil content in upland cotton using genome-wide SSRs and SNPs. Mol Breeding 39, 105 (2019). https://doi.org/10.1007/s11032-019-1007-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1007-2