Abstract
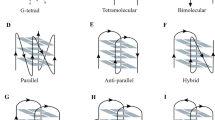
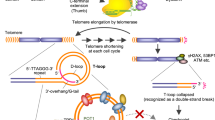
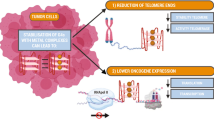
Telomeric regions contain Guanine-rich sequences arranged in a planar manner and connected by Hoogsteen hydrogen bonds that can fold into G-quadruplex (G4) DNA structures, and can be stabilized by monovalent metal cations. The presence of G4 DNA holds significance in cancer-related processes, especially due to their regulatory potential at transcriptional and translational levels of oncogene and tumor suppressor genes. The objective of this current research is to explore the evolving realm of FDA-approved protein kinase inhibitors, with a specific emphasis on their capacity to stabilize the G4 DNA structures formed at the human telomeric regions. This involves investigating the possibility of repurposing FDA-approved protein kinase inhibitors as a novel approach for targeting multiple cancer types. In this context, we have selected 16 telomeric G4 DNA structures as targets and 71 FDA-approved small-molecule protein kinase inhibitors as ligands. To investigate their binding affinities, molecular docking of human telomeric G4 DNA with nuclear protein kinase inhibitors and their corresponding co-crystalized ligands were performed. We found that Ponatinib and Lapatinib interact with all the selected G4 targets, the binding free energy calculations, and molecular dynamic simulations confirm their binding efficacy and stability. Thus, it is hypothesized that Ponatinib and Lapatinib may stabilize human telomeric G4 DNA in addition to their ability to inhibit BCR-ABL and the other members of the EGFR family. As a result, we also hypothesize that the stabilization of G4 DNA might represent an additional underlying mechanism contributing to their efficacy in exerting anti-cancer effects.
Graphical abstract
Stabilizing G4 structures at the human telomeric regions by the FDA-approved kinase inhibitors that targets nuclear kinases.






Similar content being viewed by others
Abbreviations
- G4:
-
G-quadruplex
- hTERT:
-
Human telomerase reverse transcriptase
- CML:
-
Chronic myeloid leukemia
- PDB:
-
Protein data bank
- SDF:
-
Structure data file
- RMSD:
-
Root mean square deviation
- OPLS4:
-
Optimized potential for liquid simulations
- XP:
-
Extra precision
- MMGBSA:
-
Molecular mechanics with generalized born and surface area solvation
- VGSB:
-
Variable dielectric surface generalised born
- CgenFF:
-
CHARMM general force field
- NVT:
-
Number of particles, volume, and temperature
- NPT:
-
Number of particles, pressure, and temperature
References
Phan AT, Kuryavyi V, Patel DJ (2006) DNA architecture: from G to Z. Curr Opin Struct Biol 16:288. https://doi.org/10.1016/J.SBI.2006.05.011
Wells RD (2007) Non-B DNA conformations, mutagenesis and disease. Trends Biochem Sci 32:271–278. https://doi.org/10.1016/J.TIBS.2007.04.003
Sen D, Gilbert W (1988) Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 334:364–366. https://doi.org/10.1038/334364A0
Nakanishi C, Seimiya H (2020) G-quadruplex in cancer biology and drug discovery. Biochem Biophys Res Commun 531:45–50. https://doi.org/10.1016/J.BBRC.2020.03.178
Ma Y, Iida K, Nagasawa K (2020) Topologies of G-quadruplex: biological functions and regulation by ligands. Biochem Biophys Res Commun 531:3–17. https://doi.org/10.1016/J.BBRC.2019.12.103
Largy E, Mergny JL, Gabelica V (2016) Role of alkali metal ions in G-quadruplex nucleic acid structure and stability. Met Ions Life Sci 16:203–258. https://doi.org/10.1007/978-3-319-21756-7_7
Chambers VS, Marsico G, Boutell JM, Di Antonio M, Smith GP, Balasubramanian S (2015) High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat Biotechnol 33:877–881. https://doi.org/10.1038/NBT.3295
Eddy J, Maizels N (2006) Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res 34:3887–3896. https://doi.org/10.1093/NAR/GKL529
Pavlova AV, Kubareva EA, Monakhova MV, Zvereva MI, Dolinnaya NG (2021) Impact of G-quadruplexes on the regulation of genome integrity, DNA damage and repair. Biomolecules. https://doi.org/10.3390/BIOM11091284
Balasubramanian S, Hurley LH, Neidle S (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 10:261–275. https://doi.org/10.1038/NRD3428
De S, Michor F (2011) DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat Struct Mol Biol 18:950–955. https://doi.org/10.1038/NSMB.2089
Sanchez-Martin V, Lopez-Pujante C, Soriano-Rodriguez M, Garcia-Salcedo JA (2020) An Updated focus on quadruplex structures as potential therapeutic targets in cancer. Int J Mol Sci 21:1–24. https://doi.org/10.3390/IJMS21238900
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. https://doi.org/10.1016/J.CELL.2011.02.013
Henderson E, Hardin CC, Walk SK, Tinoco I, Blackburn EH (1987) Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell 51:899–908. https://doi.org/10.1016/0092-8674(87)90577-0
Monsen RC, DeLeeuw L, Dean WL, Gray RD, Sabo TM, Chakravarthy S, Chaires JB, Trent JO (2020) The hTERT core promoter forms three parallel G-quadruplexes. Nucleic Acids Res 48:5720–5734. https://doi.org/10.1093/NAR/GKAA107
Gu W, Lin Z, Zhao S, Wang G, Shen Z, Liu W, Cai Y, Wang K, Wan CC, Yan T (2022) Research progress on G-quadruplexes in human telomeres and human telomerase reverse transcriptase (hTERT) promoter. Oxid Med Cell Longev. https://doi.org/10.1155/2022/2905663
Sanchez-Martin V, Soriano M, Garcia-Salcedo JA (2021) Quadruplex ligands in cancer therapy. Cancers (Basel). https://doi.org/10.3390/CANCERS13133156
Pirota V, Nadai M, Doria F, Richter SN (2019) Naphthalene diimides as multimodal G-quadruplex-selective ligands. Molecules. https://doi.org/10.3390/MOLECULES24030426
Burger AM, Dai F, Schultes CM, Reszka AP, Moore MJ, Double JA, Neidle S (2005) The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res 65:1489–1496. https://doi.org/10.1158/0008-5472.CAN-04-2910
Wheelhouse RT, Sun D, Han H, Han FX, Hurley LH (1998) Cationic porphyrins as telomerase inhibitors: the interaction of tetra-(N-methyl-4-pyridyl)porphine with quadruplex DNA. J Am Chem Soc 120:3261–3262. https://doi.org/10.1021/ja973792e
Liu LY, Ma TZ, Zeng YL, Liu W, Mao ZW (2022) Structural basis of pyridostatin and its derivatives specifically binding to G-quadruplexes. J Am Chem Soc 144:11878–11887. https://doi.org/10.1021/JACS.2C04775
Ghosh A, Trajkovski M, Teulade-Fichou MP, Gabelica V, Plavec J (2022) Phen-DC3 induces refolding of human telomeric dna into a chair-type antiparallel G-quadruplex through ligand intercalation. Angew Chem Int Ed Engl 5:35. https://doi.org/10.1002/ANIE.202207384
Tera M, Ishizuka H, Takagi M, Suganuma M, Shin-Ya K, Nagasawa K (2008) Macrocyclic hexaoxazoles as sequence- and mode-selective G-quadruplex binders. Angew Chem Int Ed Engl 47:5557–5560. https://doi.org/10.1002/ANIE.200801235
Tera M, Iida K, Ishizuka H, Takagi M, Suganuma M, Doi T, Shin-Ya K, Nagasawa K (2009) Synthesis of a potent G-quadruplex-binding macrocyclic heptaoxazole. ChemBioChem 10:431–435. https://doi.org/10.1002/CBIC.200800563
Rodriguez R, Miller KM, Forment JV, Bradshaw CR, Nikan M, Britton S, Oelschlaegel T, Xhemalce B, Balasubramanian S, Jackson SP (2012) Small-molecule-induced DNA damage identifies alternative DNA structures in human genes. Nat Chem Biol 8:301–310. https://doi.org/10.1038/NCHEMBIO.780
Koirala D, Dhakal S, Ashbridge B, Sannohe Y, Rodriguez R, Sugiyama H, Balasubramanian S, Mao H (2011) A single-molecule platform for investigation of interactions between G-quadruplexes and small-molecule ligands. Nat Chem 3:782–787. https://doi.org/10.1038/NCHEM.1126
Revikumar A, Kashyap V, Palollathil A, Aravind A, Raguraman R, Kumar KMK, Vijayakumar M, Prasad TSK, Raju R (2022) Multiple G-quadruplex binding ligand induced transcriptomic map of cancer cell lines. J Cell Commun Signal 16:129–135. https://doi.org/10.1007/S12079-021-00637-Z
De Lange T (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19:2100–2110. https://doi.org/10.1101/GAD.1346005
Victorelli S, Passos JF (2017) Telomeres and cell senescence—size matters not. EBioMedicine 21:14–20. https://doi.org/10.1016/J.EBIOM.2017.03.027
Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR (2007) Protein composition of catalytically active human telomerase from immortal cells. Science 315:1850–1853. https://doi.org/10.1126/SCIENCE.1138596
Trybek T, Kowalik A, Góźdź S, Kowalska A (2020) Telomeres and telomerase in oncogenesis. Oncol Lett 20:1015–1027. https://doi.org/10.3892/OL.2020.11659
Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, Wong M, Zhang Z, Li G, Sung WK, Zhou J, Chng WJ, Li S, Liu E, Tergaonkar V (2012) Telomerase directly regulates NF-κB-dependent transcription. Nat Cell Biol 14:1270–1281. https://doi.org/10.1038/NCB2621
Liu H, Liu Q, Ge Y, Zhao Q, Zheng X, Zhao Y (2016) hTERT promotes cell adhesion and migration independent of telomerase activity. Sci Rep. https://doi.org/10.1038/SREP22886
Koh CM, Khattar E, Leow SC, Liu CY, Muller J, Ang WX, Li Y, Franzoso G, Li S, Guccione E, Tergaonkar V (2015) Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J Clin Invest 125:2109–2122. https://doi.org/10.1172/JCI79134
Shay JW, Wright WE (2019) Telomeres and telomerase: three decades of progress. Nat Rev Genet 20:299–309. https://doi.org/10.1038/S41576-019-0099-1
Liu T, Yuan X, Xu D (2016) Cancer-specific telomerase reverse transcriptase (TERT) promoter mutations: biological and clinical implications. Genes (Basel). https://doi.org/10.3390/GENES7070038
Yuan X, Larsson C, Xu D (2019) Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene 38:6172–6183. https://doi.org/10.1038/S41388-019-0872-9
Tawani A, Kumar A (2015) Structural insight into the interaction of flavonoids with human telomeric sequence. Sci Rep. https://doi.org/10.1038/SREP17574
Mendes E, Aljnadi IM, Bahls B, Victor BL, Paulo A (2022) Major achievements in the design of quadruplex-interactive small molecules. Pharmaceuticals (Basel). https://doi.org/10.3390/PH15030300
Franceschin M, Rossetti L, D’Ambrosio A, Schirripa S, Bianco A, Ortaggi G, Savino M, Schultes C, Neidle S (2006) Natural and synthetic G-quadruplex interactive berberine derivatives. Bioorg Med Chem Lett 16:1707–1711. https://doi.org/10.1016/J.BMCL.2005.12.001
Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH (1997) Inhibition of human telomerase by a G-quadruplex-interactive compound. J Med Chem 40:2113–2116. https://doi.org/10.1021/JM970199Z
Zheng XH, Chen HY, Tong ML, Ji LN, Mao ZW (2012) Platinum squares with high selectivity and affinity for human telomeric G-quadruplexes. Chem Commun (Camb) 48:7607–7609. https://doi.org/10.1039/C2CC32942E
Tawani A, Amanullah A, Mishra A, Kumar A (2016) Evidences for piperine inhibiting cancer by targeting human G-quadruplex DNA sequences. Sci Rep. https://doi.org/10.1038/SREP39239
Tyagi S, Saxena S, Srivastava P, Sharma T, Kundu N, Kaur S, Shankaraswamy J (2020) Screening the binding potential of quercetin with parallel, antiparallel and mixed G-quadruplexes of human telomere and cancer protooncogenes using molecular docking approach. SN Appl Sci 2:1–16. https://doi.org/10.1007/S42452-020-2280-8/TABLES/3
Jha NS, Mishra S, Mamidi AS, Mishra A, Jha SK, Surolia A (2016) Targeting human telomeric G-quadruplex DNA with curcumin and its synthesized analogues under molecular crowding conditions. RSC Adv 6:7474–7487. https://doi.org/10.1039/C5RA17390F
Revikumar A (2021) Amentoflavone as a potential telomeric G-quadruplex DNA stabilizing agent. Gene Rep 22:100976. https://doi.org/10.1016/j.genrep.2020.100976
Fabbro D, Cowan-Jacob SW, Moebitz H (2015) Ten things you should know about protein kinases: IUPHAR review 14. Br J Pharmacol 172:2675–2700. https://doi.org/10.1111/BPH.13096
Iqbal N, Iqbal N (2014) Imatinib: a breakthrough of targeted therapy in cancer. Chemother Res Pract 2014:1–9. https://doi.org/10.1155/2014/357027
Kancha RK, Von Bubnoff N, Miething C, Peschel C, Götze KS, Duyster J (2008) Imatinib and leptomycin B are effective in overcoming imatinib-resistance due to Bcr-Abl amplification and clonal evolution but not due to Bcr-Abl kinase domain mutation. Haematologica 93:1718–1722. https://doi.org/10.3324/HAEMATOL.13207
Wang YH, Yang QF, Lin X, Chen D, Wang ZY, Chen B, Han HY, Di CH, Cai KC, Li Q, Yang S, Tang YL, Li F (2022) G4LDB 2.2: a database for discovering and studying G-quadruplex and i-motif ligands. Nucleic Acids Res 50:D150–D160. https://doi.org/10.1093/NAR/GKAB952
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242. https://doi.org/10.1093/NAR/28.1.235
Roskoski R (2023) Properties of FDA-approved small molecule protein kinase inhibitors: a 2023 update. Pharmacol Res. https://doi.org/10.1016/J.PHRS.2022.106552
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, Wang J, Yu B, Zhang J, Bryant SH (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213. https://doi.org/10.1093/NAR/GKV951
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminform. https://doi.org/10.1186/1758-2946-3-33
Reimand J, Kull M, Peterson H, Hansen J, Vilo J (2007) g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. https://doi.org/10.1093/NAR/GKM226
Madhavi Sastry G, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/S10822-013-9644-8
Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT (2006) Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49:6177–6196. https://doi.org/10.1021/JM051256O
Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE (2021) UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30:70–82. https://doi.org/10.1002/PRO.3943
Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all-atom protein loop prediction. Proteins 55:351–367. https://doi.org/10.1002/PROT.10613
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindah E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/J.SOFTX.2015.06.001
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Hart K, Foloppe N, Baker CM, Denning EJ, Nilsson L, MacKerell AD (2012) Optimization of the CHARMM additive force field for DNA: improved treatment of the BI/BII conformational equilibrium. J Chem Theory Comput 8:348–362. https://doi.org/10.1021/CT200723Y
Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, De Groot BL, Grubmüller H, MacKerell AD (2017) CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14:71–73. https://doi.org/10.1038/NMETH.4067
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690. https://doi.org/10.1002/JCC.21367
Sun D, Lakkaraju SK, Jo S, Mackerell AD (2018) Determination of ionic hydration free energies with grand canonical Monte Carlo/molecular dynamics simulations in explicit water. J Chem Theory Comput 14:5290–5302. https://doi.org/10.1021/ACS.JCTC.8B00604
Mauri M, Elli T, Caviglia G, Uboldi G, Azzi M (2017) RAWGraphs: a visualisation platform to create open outputs. ACM International Conference Proceeding Series Part F131371. https://doi.org/10.1145/3125571.3125585
de Lange T, Shiue L, Myers RM, Cox DR, Naylor SL, Killery AM, Varmus HE (1990) Structure and variability of human chromosome ends. Mol Cell Biol 10:518–527. https://doi.org/10.1128/MCB.10.2.518-527.1990
Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH (2002) Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular g-quadruplex. J Am Chem Soc 124:2098–2099. https://doi.org/10.1021/JA017308Q
Cortes JE, Kantarjian H, Shah NP, Bixby D, Mauro MJ, Flinn I, O’Hare T, Hu S, Narasimhan NI, Rivera VM, Clackson T, Turner CD, Haluska FG, Druker BJ, Deininger MWN, Talpaz M (2012) Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med 367:2075–2088. https://doi.org/10.1056/NEJMOA1205127
Sanford D, Kantarjtan H, Skinner J, Jabbour E, Cortes J (2015) Phase II trial of ponatinib in patients with chronic myeloid leukemia resistant to one previous tyrosine kinase inhibitor. Haematologica 100:e494–e495. https://doi.org/10.3324/HAEMATOL.2015.132845
Cortes JE, Kim D-W, Pinilla-Ibarz J, le Coutre P, Paquette R, Chuah C, Nicolini FE, Apperley JF, Khoury HJ, Talpaz M, DiPersio J, DeAngelo DJ, Abruzzese E, Rea D, Baccarani M, Müller MC, Gambacorti-Passerini C, Wong S, Lustgarten S, Rivera VM, Clackson T, Turner CD, Haluska FG, Guilhot F, Deininger MW, Hochhaus A, Hughes T, Goldman JM, Shah NP, Kantarjian H (2013) A phase 2 trial of ponatinib in Philadelphia chromosome-positive leukemias. N Engl J Med 369:1783–1796. https://doi.org/10.1056/NEJMOA1306494
Tan FH, Putoczki TL, Stylli SS, Luwor RB (2019) Ponatinib: a novel multi-tyrosine kinase inhibitor against human malignancies. Onco Targets Ther 12:635–645. https://doi.org/10.2147/OTT.S189391
The characterization of novel, dual ErbB-2/EGFR, tyrosine kinase inhibitors: potential therapy for cancer-PubMed. https://pubmed.ncbi.nlm.nih.gov/11585755/. Accessed 26 Aug 2023
Salem AA, El Haty IA, Ghattas MA (2022) GW-2974 and SCH-442416 modulators of tyrosine kinase and adenosine receptors can also stabilize human telomeric G-quadruplex DNA. PLoS ONE. https://doi.org/10.1371/JOURNAL.PONE.0277963
Acknowledgements
We thank the Yenepoya (Deemed to be University) for the facility support to the Centre for Integrative Omics Data Science (CIODS). Amjesh Revikumar is a recipient of a national post-doctoral fellowship (PDF/2017/001652) and Rajesh Raju is a recipient of the Young Scientist Award (YSS/2014/000607) from the Science and Engineering Research Board, Department of Science and Technology (DST), Government of India.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
AR: developed the original hypothesis for the work. AR and BB: developed the computational workflow and wrote the initial draft of the manuscript. BB, AJK, and HK: did the Molecular docking and dynamics studies. LJ: developed the graphical contents in the manuscript. SS, RR: critically evaluated the manuscript and ensured that the study’s scientific aspect was rationally valid. All authors read, edited, and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Banjan, B., Koshy, A.J., Kalath, H. et al. Potential protein kinase inhibitors that target G-quadruplex DNA structures in the human telomeric regions. Mol Divers (2024). https://doi.org/10.1007/s11030-023-10768-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10768-7