Abstract
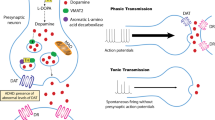
The dopamine transporter (DAT), responsible for the regulation of dopaminergic neurotransmission, is implicated in the etiology of several neuropsychiatric disorders which, in turn, have contributed to high rates of disability and numerous deaths in recent years, significantly impacting the global health system. Although the research for new drugs for the treatment of neuropsychiatric disorders has evolved in recent years, the availability of DAT-selective drugs that do not generate the same psychostimulant effects observed in drugs of abuse remains scarce. Therefore, we performed a QSAR study based on a dataset of 36 methylamine derivatives described as DAT inhibitors. The model was obtained based only in descriptors derived from 2D structures, and it was validated and generated satisfactory results considering the metrics used for internal and external validation. Subsequently, a virtual screening step also based on 2D similarity was performed, where it was possible to identify a total of 1157 compounds. After a series of reductions of the set using toxicity filters, applicability domain evaluation, and pharmacokinetic properties in silico assessment, seven hit compounds were selected as the most promising to be used, in future studies, as new scaffolds for the development of new DAT inhibitors.










Similar content being viewed by others
References
Merikangas AK, Almasy L (2020) Using the tools of genetic epidemiology to understand sex differences in neuropsychiatric disorders. Genes Brain Behav 19:e12660. https://doi.org/10.1111/gbb.12660
Williams OOF, Coppolino M, George SR et al (2021) Sex differences in dopamine receptors and relevance to neuropsychiatric disorders. Brain Sci 11:1199. https://doi.org/10.3390/brainsci11091199
Chen R, Ferris MJ, Wang S (2020) Dopamine D2 autoreceptor interactome: targeting the receptor complex as a strategy for treatment of substance use disorder. Pharmacol Ther 213:107583. https://doi.org/10.1016/j.pharmthera.2020.107583
Krout D, Pramod AB, Dahal RA et al (2017) Inhibitor mechanisms in the S1 binding site of the dopamine transporter defined by multi-site molecular tethering of photoactive cocaine analogs. Biochem Pharmacol 142:204–215. https://doi.org/10.1016/j.bcp.2017.07.015
Ali SW, Pereira F (2017) Dopamine: neuropsychiatric disorders and neurotoxicity. Toxicol Lett 280:S62. https://doi.org/10.1016/j.toxlet.2017.07.156
Torres GE, Gainetdinov RR, Caron MG (2003) Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci 4:13–25. https://doi.org/10.1038/nrn1008
Reith MEA, Blough BE, Hong WC et al (2015) Behavioral, biological, and chemical perspectives on atypical agents targeting the dopamine transporter. Drug Alcohol Depend 147:1–19. https://doi.org/10.1016/j.drugalcdep.2014.12.005
Stępnicki P, Kondej M, Kaczor AA (2018) Current concepts and treatments of schizophrenia. Molecules 23:2087. https://doi.org/10.3390/molecules23082087
Baig MH, Ahmad K, Rabbani G et al (2017) Computer aided drug design and its application to the development of potential drugs for neurodegenerative disorders. Curr Neuropharmacol 16:740–748. https://doi.org/10.2174/1570159x15666171016163510
Carmo Bastos ML, Silva-Silva JV, Cruz JN et al (2023) Alkaloid from Geissospermum sericeum Benth. & Hook.f. ex Miers (Apocynaceae) induce apoptosis by caspase pathway in human gastric cancer cells. Pharmaceuticals 16:765. https://doi.org/10.3390/ph16050765
da Silva DF, Souza JL, Costa DM et al (2023) Antiplasmodial activity of coumarins isolated from Polygala boliviensis: in vitro and in silico studies. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2023.2173295
Almeida VM, Dias ÊR, Souza BC et al (2022) Methoxylated flavonols from Vellozia dasypus Seub ethyl acetate active myeloperoxidase extract: in vitro and in silico assays. J Biomol Struct Dyn 40:7574–7583. https://doi.org/10.1080/07391102.2021.1900916
Rego CMA, Francisco AF, Boeno CN et al (2022) Inflammasome NLRP3 activation induced by Convulxin, a C-type lectin-like isolated from Crotalus durissus terrificus snake venom. Sci Rep 12:1–17. https://doi.org/10.1038/s41598-022-08735-7
Santos CBR, Santos KLB, Cruz JN et al (2021) Molecular modeling approaches of selective adenosine receptor type 2A agonists as potential anti-inflammatory drugs. J Biomol Struct Dyn 39:3115–3127. https://doi.org/10.1080/07391102.2020.1761878
Lima M, Siqueira AS, Möller MLS et al (2022) In silico improvement of the cyanobacterial lectin microvirin and mannose interaction. J Biomol Struct Dyn 40:1064–1073. https://doi.org/10.1080/07391102.2020.1821782
Santos KLB, Cruz JN, Silva LB et al (2020) Identification of novel chemical entities for adenosine receptor type 2a using molecular modeling approaches. Molecules 25:1245. https://doi.org/10.3390/molecules25051245
Silva LB, Ferreira EFB, Satti MR et al (2023) Galantamine based novel acetylcholinesterase enzyme inhibitors: a molecular modeling design approach. Molecules 28:1035. https://doi.org/10.3390/molecules28031035
El Fadili M, Er-Rajy M, Kara M et al (2022) QSAR, ADMET in silico pharmacokinetics, molecular docking and molecular dynamics studies of novel bicyclo (aryl methyl) benzamides as potent GlyT1 inhibitors for the treatment of schizophrenia. Pharmaceuticals 15:670. https://doi.org/10.3390/ph15060670
El fadili M, Er-rajy M, Imtara H et al (2023) QSAR, ADME-Tox, molecular docking and molecular dynamics simulations of novel selective glycine transporter type 1 inhibitors with memory enhancing properties. Heliyon 9:e13706. https://doi.org/10.1016/j.heliyon.2023.e13706
Almeida RBM, Barbosa DB, Bomfim MR et al (2023) Identification of a novel dual inhibitor of acetylcholinesterase and butyrylcholinesterase: in vitro and in silico studies. Pharmaceuticals 16:95. https://doi.org/10.3390/ph16010095
Shao L, Hewitt MC, Wang F et al (2011) Discovery of N-methyl-1-(1-phenylcyclohexyl)methanamine, a novel triple serotonin, norepinephrine, and dopamine reuptake inhibitor. Bioorg Med Chem Lett 21:1438–1441. https://doi.org/10.1016/j.bmcl.2011.01.016
Macêdo WJC, Braga FS, Santos CF et al (2015) Antimalarial artemisinins derivatives study: molecular modeling and multivariate analysis (PCA, HCA, KNN, SIMCA and SDA). J Comput Theor Nanosci 12:3443–3458. https://doi.org/10.1166/jctn.2015.4138
Vieira JB, Braga FS, Lobato CC et al (2014) A QSAR, pharmacokinetic and toxicological study of new artemisinin compounds with anticancer activity. Molecules 19:10670–10697. https://doi.org/10.3390/molecules190810670
ACD/Chemsketch Freeware (2023) https://www.acdlabs.com. Accessed 23 Aug 2023
Weininger D (1988) SMILES, a chemical language and information system: 1: introduction to methodology and encoding rules. J Chem Inform Comput Sci 28:31–36. https://doi.org/10.1021/ci00057a005
Helguera A, Combes R, Gonzalez M et al (2008) Applications of 2D descriptors in drug design: a DRAGON tale. Curr Top Med Chem 8:1628–1655. https://doi.org/10.2174/156802608786786598
Roque JV, Cardoso W, Peternelli LA et al (2019) Comprehensive new approaches for variable selection using ordered predictors selection. Anal Chim Acta 1075:57–70. https://doi.org/10.1016/j.aca.2019.05.039
Ferreira MMC, Montanari CA, Gaudio AC (2002) Variable selection in QSAR. Quim Nova 25:439–448. https://doi.org/10.1590/s0100-40422002000300017
Roy K, Kar S, Das RN (2015) Classical QSAR. Understanding the basics of qsar for applications in pharmaceutical sciences and risk assessment, 1st edn. Academic Press, London, pp 81–102
Gaudio AC, Zandonade E (2001) Proposition, validation and analysis of QSAR models. Quim Nova 24:658–671. https://doi.org/10.1590/s0100-40422001000500013
Galvão RKH, Araújo MCU, Silva EC et al (2007) Cross-validation for the selection of spectral variables using the successive projections algorithm. J Braz Chem Soc 18:1580–1584. https://doi.org/10.1590/S0103-50532007000800021
Roy PP, Paul S, Mitra I et al (2009) On two novel parameters for validation of predictive QSAR models. Molecules 14:1660–1701. https://doi.org/10.3390/molecules14051660
Kiralj R, Ferreira MMC (2009) Basic validation procedures for regression models in QSAR and QSPR studies: theory and application. J Braz Chem Soc 20:770–787. https://doi.org/10.1590/S0103-50532009000400021
Martins JPA, Ferreira MMC (2013) QSAR modeling: um novo pacote computacional open source para gerar e validar modelos QSAR. Quim Nova 36:554–560. https://doi.org/10.1590/s0100-40422013000400013
Roy K, Das RN, Ambure P et al (2016) Be aware of error measures. Further studies on validation of predictive QSAR models. Chemom Intell Lab Syst 152:18–33. https://doi.org/10.1016/j.chemolab.2016.01.008
Roy K, Mitra I (2012) On the use of the metric rm2 as an effective tool for validation of QSAR models in computational drug design and predictive toxicology. Mini-Rev Med Chem 12:491–504. https://doi.org/10.2174/138955712800493861
Chirico N, Gramatica P (2011) Real external predictivity of QSAR models: how to evaluate it? Comparison of different validation criteria and proposal of using the concordance correlation coefficient. J Chem Inform Model 51:2320–2335. https://doi.org/10.1021/ci200211n
Tropsha PG, Gombar VK (2003) The importance of being earnest: Validation is the absolute essential for successful application and interpretation of QSPR models, QSAR Comb. Sci 22:69–77. https://doi.org/10.1002/qsar.200390007
Roy K, Kar S (2014) The rm2 metrics and regression through origin approach: reliable and useful validation tools for predictive QSAR models (Commentary on ’Is regression through origin useful in external validation of QSAR models?’). Eur J Pharm Sci 62:111–114. https://doi.org/10.1016/j.ejps.2014.05.019
Roy K, Kar S, Ambure P (2015) On a simple approach for determining applicability domain of QSAR models. Chemom Intell Lab Syst 145:22–29. https://doi.org/10.1016/j.chemolab.2015.04.013
Jaworska J, Nikolova-Jeliazkova N, Aldenberg T (2005) QSAR applicability domain estimation by projection of the training set in descriptor space: a review. ATLA Altern Lab Anim 33:445–459. https://doi.org/10.1177/026119290503300508
Minovski N, Župerl Š, Drgan V et al (2013) Assessment of applicability domain for multivariate counter-propagation artificial neural network predictive models by minimum Euclidean distance space analysis: a case study. Anal Chim Acta 759:28–42. https://doi.org/10.1016/j.aca.2012.11.002
Bocci G, Carosati E, Vayer P et al (2017) ADME-Space: a new tool for medicinal chemists to explore ADME properties. Sci Rep 7:6359. https://doi.org/10.1038/s41598-017-06692-0
Daina A, Zoete V (2016) A BOILED-Egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem 11:1117–1121. https://doi.org/10.1002/cmdc.201600182
Jeynes A, Provias J (2011) An investigation into the role of P-glycoprotein in Alzheimer’s disease lesion pathogenesis. Neurosci Lett 487:389–393. https://doi.org/10.1016/j.neulet.2010.10.063
Seo M, Chae CH, Lee Y et al (2021) Novel QSAR models for molecular initiating event modeling in two intersecting adverse outcome pathways based pulmonary fibrosis prediction for biocidal mixtures. Toxics 9:59. https://doi.org/10.3390/toxics9030059
Greenland S, Senn SJ, Rothman KJ et al (2016) Statistical tests, P values, confidence intervals, and power: a guide to misinterpretations. Eur J Epidemiol 31:337–350. https://doi.org/10.1007/s10654-016-0149-3
Liu P, Long W (2009) Current mathematical methods used in QSAR/QSPR studies. Int J Mol Sci 10:1978–1998. https://doi.org/10.3390/ijms10051978
Čampulová M, Michálek J, Mikuška P et al (2018) Nonparametric algorithm for identification of outliers in environmental data. J Chemom 32:e2997. https://doi.org/10.1002/cem.2997
Yosipof A, Senderowitz H (2015) K-Nearest neighbors optimization-based outlier removal. J Comput Chem 36:493–506. https://doi.org/10.1002/jcc.23803
Cao DS, Deng ZK, Zhu MF et al (2017) Ensemble partial least squares regression for descriptor selection, outlier detection, applicability domain assessment, and ensemble modeling in QSAR/QSPR modeling. J Chemom 31:e2922. https://doi.org/10.1002/cem.2922
Kaneko H (2019) Estimation of predictive performance for test data in applicability domains using y-randomization. J Chemom 33:e3171. https://doi.org/10.1002/cem.3171
Shayanfar A, Shayanfar S (2014) Is regression through origin useful in external validation of QSAR models? Eur J Pharm Sci 59:31–35. https://doi.org/10.1016/j.ejps.2014.03.007
Majumdar S, Basak SC (2018) Beware of external validation!—a comparative study of several validation techniques used in QSAR modelling. Curr Comp Aided Drug Des 14:284–291. https://doi.org/10.2174/1573409914666180426144304
Gramatica P, Sangion A (2016) A historical excursus on the statistical validation parameters for QSAR Models: a clarification concerning metrics and terminology. J Chem Inform Model 56:1127–1131. https://doi.org/10.1021/acs.jcim.6b00088
Kubinyi H, Hamprecht FA, Mietzner T (1998) Three-dimensional quantitative similarity-activity relationships (3D QSiAR) from SEAL similarity matrices. J Med Chem 41:2553–2564. https://doi.org/10.1021/jm970732a
Olasupo SB, Uzairu A, Shallangwa GA et al (2020) Chemoinformatic studies on some inhibitors of dopamine transporter and the receptor targeting schizophrenia for developing novel antipsychotic agents. Heliyon 6:e04464. https://doi.org/10.1016/j.heliyon.2020.e04464
Geppert H, Vogt M, Bajorath J (2010) Current trends in ligand-based virtual screening: molecular representations, data mining methods, new application areas, and performance evaluation. J Chem Inform Model 50:205–216. https://doi.org/10.1021/ci900419k
Neves BJ, Braga RC, Melo-Filho CC et al (2018) QSAR-based virtual screening: advances and applications in drug discovery. Front Pharmacol 9:1275. https://doi.org/10.3389/fphar.2018.01275
Grisoni F, Consonni V, Todeschini R (2018) Impact of molecular descriptors on computational models. Methods Mol Biol 1825:171–209. https://doi.org/10.1007/978-1-4939-8639-2_5
Liu R, Wallqvist A (2019) Molecular similarity-based domain applicability metric efficiently identifies out-of-domain compounds. J Chem Inform Model 59:181–189. https://doi.org/10.1021/acs.jcim.8b00597
Berenger F, Yamanishi Y (2019) A distance-based Boolean applicability domain for classification of high throughput screening data. J Chem Inform Model 59:463–476. https://doi.org/10.1021/acs.jcim.8b00499
Pantaleão SQ, Fernandes PO, Gonçalves JE et al (2022) Recent advances in the prediction of pharmacokinetics properties in drug design studies: a review. ChemMedChem 17:e202100542. https://doi.org/10.1002/cmdc.202100542
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Tomlinson MJ, Krout D, Pramod AB et al (2019) Identification of the benztropine analog [125I]GA II 34 binding site on the human dopamine transporter. Neurochem Int 123:34–45. https://doi.org/10.1016/j.neuint.2018.08.008
Wang J, Hou T (2015) Advances in computationally modeling human oral bioavailability. Adv Drug Deliv Rev 86:11–16. https://doi.org/10.1016/j.addr.2015.01.001
Lipinski CA, Lombardo F, Dominy BW et al (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26. https://doi.org/10.1016/S0169-409X(00)00129-0
Veber DF, Johnson SR, Cheng HY et al (2002) Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 45:2615–2623. https://doi.org/10.1021/jm020017n
Ghose AK, Viswanadhan VN, Wendoloski JJ (1999) A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J Comb Chem 1:55–68. https://doi.org/10.1021/cc9800071
Egan WJ, Merz KM, Baldwin JJ (2000) Prediction of drug absorption using multivariate statistics. J Med Chem 43:3867–3877. https://doi.org/10.1021/jm000292e
Muegge I, Heald SL, Brittelli D (2001) Simple selection criteria for drug-like chemical matter. J Med Chem 44:1841–1846. https://doi.org/10.1021/jm015507e
Ertl P, Rohde B, Selzer P (2000) Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem 43:3714–3717. https://doi.org/10.1021/jm000942e
Prasanna S, Doerksen R (2008) Topological polar surface area: a useful descriptor in 2D-QSAR. Curr Med Chem 16:21–41. https://doi.org/10.2174/092986709787002817
Abuhassan Q, Khadra I, Pyper K et al (2022) Structured solubility behaviour in bioequivalent fasted simulated intestinal fluids. Eur J Pharm Biopharm 176:108–121. https://doi.org/10.1016/j.ejpb.2022.05.010
Delaney JS (2004) ESOL: estimating aqueous solubility directly from molecular structure. J Chem Inform Comput Sci 44:1000–1005. https://doi.org/10.1021/ci034243x
Ali J, Camilleri P, Brown MB et al (2012) Revisiting the general solubility equation: in silico prediction of aqueous solubility incorporating the effect of topographical polar surface area. J Chem Inform Model 52:420–428. https://doi.org/10.1021/ci200387c
Dahan A, Miller JM (2012) The solubility-permeability interplay and its implications in formulation design and development for poorly soluble drugs. AAPS J 14:244–251. https://doi.org/10.1208/s12248-012-9337-6
Keaney J, Campbell M (2015) The dynamic blood-brain barrier. FEBS J 282:4067–4079. https://doi.org/10.1111/febs.13412
Poongavanam V, Haider N, Ecker GF (2012) Fingerprint-based in silico models for the prediction of P-glycoprotein substrates and inhibitors. Bioorg Med Chem 20:5388–5395. https://doi.org/10.1016/j.bmc.2012.03.045
Cummins CL, Salphati L, Reid MJ et al (2003) In vivo modulation of intestinal CYP3A metabolism by P-glycoprotein: studies using the rat single-pass intestinal perfusion model. J Pharmacol Exp Ther 305:306–314. https://doi.org/10.1124/jpet.102.044719
Saaby L, Brodin B (2017) A critical view on in vitro analysis of P-glycoprotein (P-gp) transport kinetics. J Pharm Sci 106:2257–2264. https://doi.org/10.1016/j.xphs.2017.04.022
Terasaki T (2017) Quantitative expression of ADME proteins at the blood-brain barrier. Drug Metab Pharmacokinet 32:S12. https://doi.org/10.1016/j.dmpk.2016.10.059
Manikandan P, Nagini S (2017) Cytochrome P450 structure, function and clinical significance: a review. Curr Drug Targets 19:38–54. https://doi.org/10.2174/1389450118666170125144557
Zanger UM, Schwab M (2013) Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 138:103–141. https://doi.org/10.1016/j.pharmthera.2012.12.007
Zhou SF (2009) Polymorphism of human cytochrome P450 2D6 and its clinical significance: part II. Clin Pharmacokinet 48:761–804. https://doi.org/10.2165/11318070-000000000-00000
Thurkauf A, Costa B, Yamaguchi S et al (1990) Synthesis and anticonvulsant activity of 1-phenylcyclohexylamine analogs. J Med Chem 33:1452–1458. https://doi.org/10.1021/jm00167a027
Acknowledgements
The authors would like to thank the Fundação Araucária (FAPPR) and PROAP/CAPES.
Funding
Fundação Araucária.
Author information
Authors and Affiliations
Contributions
All authors contributed equally to the study, through periodic meetings to discuss the study data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors have no relevant financial or nonfinancial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
de Oliveira, L.H.D., Cruz, J.N., dos Santos, C.B.R. et al. Multivariate QSAR, similarity search and ADMET studies based in a set of methylamine derivatives described as dopamine transporter inhibitors. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10724-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10724-5