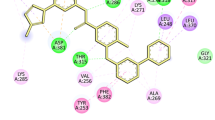
Abstract
Parkinson’s disease is characterized by a multifactorial nature that is linked to different pathways. Among them, the abnormal deposition and accumulation of α-synuclein fibrils is considered a neuropathological hallmark of Parkinson’s disease. Several synthetic and natural compounds have been tested for their potency to inhibit the aggregation of α-synuclein. However, the molecular mechanisms responsible for the potency of these drugs to further rationalize their development and optimization are yet to be determined. To enhance our understanding of the structural requirements necessary for modulating the aggregation of α-synuclein fibrils, we retrieved a large dataset of α-synuclein inhibitors with their reported potency from the ChEMBL database to explore their chemical space and to generate QSAR models for predicting new bioactive compounds. The best performing QSAR model was applied to the LOTUS natural products database to screen for potential α-synuclein inhibitors followed by a pharmacophore design using the representative compounds sampled from each cluster in the ChEMBL dataset. Five natural products were retained after molecular docking studies displaying a binding affinity of − 6.0 kcal/mol or lower. ADMET analysis revealed satisfactory properties and predicted that all the compounds can cross the blood–brain barrier and reach their target. Finally, molecular dynamics simulations demonstrated the superior stability of LTS0078917 compared to the clinical candidate, Anle138b. We found that LTS0078917 shows promise in stabilizing the α-synuclein monomer by specifically binding to its hairpin-like coil within the N-terminal region. Our dynamic analysis of the inhibitor-monomer complex revealed a tendency towards a more compact conformation, potentially reducing the likelihood of adopting an elongated structure that favors the formation and aggregation of pathological oligomers. These findings offer valuable insights for the development of novel α-synuclein inhibitors derived from natural sources.
Graphical abstract
















Similar content being viewed by others
References
Dugger BN, Dickson DW (2017) Pathology of neurodegenerative diseases. Cold Spring Harb Perspect Biol 9(7):a028035
Hardy J, Gwinn-Hardy K (1998) Genetic classification of primary neurodegenerative disease. Science 282(5391):1075–1079
Goetz CG, Emre M, Dubois B (2008) Parkinson’s disease dementia: definitions, guidelines, and research perspectives in diagnosis. Ann Neurol 64(S2):S81–S92
Dorsey EA, Constantinescu R, Thompson JP, Biglan KM, Holloway RG, Kieburtz K, Tanner CM (2007) Projected number of people with Parkinson disease in the most populous nations, 2005 through 2030. Neurology 68(5):384–386
Wanneveich M, Moisan F, Jacqmin-Gadda H, Elbaz A, Joly P (2018) Projections of prevalence, lifetime risk, and life expectancy of Parkinson’s disease (2010–2030) in France. Mov Disord 33(9):1449–1455
Martinez-Martin P, Rodriguez-Blazquez C, Paz S, Forjaz MJ, Frades-Payo B, Cubo E, ELEP Group (2015) Parkinson symptoms and health related quality of life as predictors of costs: a longitudinal observational study with linear mixed model analysis. PLoS ONE 10(12):e0145310
Chaudhuri KR, Healy DG, Schapira AH (2006) Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol 5(3):235–245
Jenner P (2003) Oxidative stress in Parkinson’s disease. Ann Neurol 53(S3):S26–S38
Kalia LV, Kalia SK, Lang AE (2015) Disease-modifying strategies for Parkinson’s disease. Mov Disord 30(11):1442–1450
Olanow CW, Agid Y, Mizuno Y, Albanese A, Bonucelli U, Damier P, Stocchi F (2004) Levodopa in the treatment of Parkinson’s disease: current controversies. Mov Disord 19(9):997–1005
Finberg JP (2019) Inhibitors of MAO-B and COMT: their effects on brain dopamine levels and uses in Parkinson’s disease. J Neural Transm 126(4):433–448
Szabo N, Kincses ZT, Vecsei L (2011) Novel therapy in Parkinson’s disease: adenosine A2A receptor antagonists. Expert Opin Drug Metab Toxicol 7(4):441–455
Vanle B, Olcott W, Jimenez J, Bashmi L, Danovitch I, IsHak WW (2018) NMDA antagonists for treating the non-motor symptoms in Parkinson’s disease. Transl Psychiatry 8(1):1–15
Amer DA, Irvine GB, El-Agnaf O (2006) Inhibitors of α-synuclein oligomerization and toxicity: a future therapeutic strategy for Parkinson’s disease and related disorders. Exp Brain Res 173(2):223–233
Rangasamy SB, Soderstrom K, Bakay RA, Kordower JH (2010) Neurotrophic factor therapy for Parkinson’s disease. Prog Brain Res 184:237–264
Boulaamane Y, Ibrahim MA, Britel MR, Maurady A (2022) In silico studies of natural product-like caffeine derivatives as potential MAO-B inhibitors/AA2AR antagonists for the treatment of Parkinson’s disease. J Integr Bioinform. 19(4):20210027
Oliveira L, Gasser T, Edwards R, Zweckstetter M, Melki R, Stefanis L, Outeiro TF (2021) Alpha-synuclein research: defining strategic moves in the battle against Parkinson’s disease. Npj Parkinson’s Dis 7(1):1–23
Das S, Pukala TL, Smid SD (2018) Exploring the structural diversity in inhibitors of α-synuclein amyloidogenic folding, aggregation, and neurotoxicity. Front Chem 6:181
Cheng F, Vivacqua G, Yu S (2011) The role of alpha-synuclein in neurotransmission and synaptic plasticity. J Chem Neuroanat 42(4):242–248
Takeda A, Mallory M, Sundsmo M, Honer W, Hansen L, Masliah E (1998) Abnormal accumulation of NACP/alpha-synuclein in neurodegenerative disorders. Am J Pathol 152(2):367
Gibb WR, Lees A (1988) The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J Neurol Neurosurg Psychiatry 51(6):745–752
Vicario M, Cieri D, Brini M, Calì T (2018) The close encounter between alpha-synuclein and mitochondria. Front Neurosci 12:388
Wagner J, Ryazanov S, Leonov A, Levin J, Shi S, Schmidt F, Giese A (2013) Anle138b: a novel oligomer modulator for disease-modifying therapy of neurodegenerative diseases such as prion and Parkinson’s disease. Acta Neuropathologica 125(6):795–813
Price DL, Koike MA, Khan A, Wrasidlo W, Rockenstein E, Masliah E, Bonhaus D (2018) The small molecule alpha-synuclein misfolding inhibitor, NPT200-11, produces multiple benefits in an animal model of Parkinson’s disease. Sci Rep 8(1):1–12
Ulmer TS, Bax A, Cole NB, Nussbaum RL (2005) Structure and dynamics of micelle-bound human α-synuclein. J Biol Chem 280(10):9595–9603
Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, Lindquist S (2006) α-Synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science 313(5785):324–328
Amos SBT, Schwarz TC, Shi J, Cossins BP, Baker TS, Taylor RJ, Sansom MS (2021) Membrane interactions of α-Synuclein revealed by multiscale molecular dynamics simulations, Markov state models, and NMR. J Phys Chem B 125(11):2929–2941
Kondratyev MS, Rudnev VR, Nikolsky KS, Petrovsky DV, Kulikova LI, Malsagova KA, Kaysheva AL (2022) In silico study of the interactions of anle138b isomer, an inhibitor of amyloid aggregation, with partner proteins. Int J Mol Sci 23(24):16096
Baul HS, Rajiniraja M (2018) Favorable binding of quercetin to α-synuclein as potential target in Parkinson disease: an insilico approach. Res J Pharm Technol 11(1):203–206
Zhu M, Han S, Fink AL (2013) Oxidized quercetin inhibits α-synuclein fibrillization. Biochimica et Biophysica Acta (BBA) 1830(4):2872–2881
Ahn TB, Jeon BS (2015) The role of quercetin on the survival of neuron-like PC12 cells and the expression of α-synuclein. Neural Regen Res 10(7):1113
Bisi N, Feni L, Peqini K, Pérez-Peña H, Ongeri S, Pieraccini S, Pellegrino S (2021) α-Synuclein: an all-inclusive trip around its structure, influencing factors and applied techniques. Front Chem 9:666585
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Overington JP (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic acids Res 40(D1):D1100–D1107
Bajusz D, Rácz A, Héberger K (2015) Why is Tanimoto index an appropriate choice for fingerprint-based similarity calculations? J Cheminform 7(1):1–13
Landrum, G. (2013). RDKit: a software suite for cheminformatics, computational chemistry, and predictive modeling. Greg Landrum
Capecchi A, Probst D, Reymond JL (2020) One molecular fingerprint to rule them all: drugs, biomolecules, and the metabolome. J Cheminform 12(1):1–15
Rutz A, Sorokina M, Galgonek J, Mietchen D, Willighagen E, Gaudry A, Allard PM (2022) The LOTUS initiative for open knowledge management in natural products research. elife 11:e70780
Lipinski CA (2004) Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov Today Technol 1(4):337–341
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 55(2):460–473
Sunseri J, Koes DR (2016) Pharmit: interactive exploration of chemical space. Nucleic Acids Res 44(W1):W442–W448
Huey R, Morris GM, Forli S (2012) Using AutoDock 4 and AutoDock vina with AutoDockTools: a tutorial. Scripps Res Inst Mol Graphics Lab 10550:92037
Volkamer A, Griewel A, Grombacher T, Rarey M (2010) Analyzing the topology of active sites: on the prediction of pockets and subpockets. J Chem Inf Model 50(11):2041–2052
Volkamer A, Kuhn D, Grombacher T, Rippmann F, Rarey M (2012) Combining global and local measures for structure-based druggability predictions. J Chem Inf Model 52(2):360–372
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Bhakhar KA, Gajjar ND, Bodiwala KB, Sureja DK, Dhameliya TM (2021) Identification of anti-mycobacterial agents against mmpL3: virtual screening, ADMET analysis and MD simulations. J Mol Struct 1244:130941
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M (2015) PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res 43(W1):W443–W447
Norinder U, Bergström CA (2006) Prediction of ADMET properties. Chem Enab Drug Discov 1(9):920–937
Cheng F, Li W, Liu G, Tang Y (2013) In silico ADMET prediction: recent advances, current challenges and future trends. Curr Top Med Chem 13(11):1273–1289
Rao VS, Srinivas K (2011) Modern drug discovery process: an in silico approach. J Bioinform Sequence Anal 2(5):89–94
Pires DE, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58(9):4066–4072
Sureja DK, Shah AP, Gajjar ND, Jadeja SB, Bodiwala KB, Dhameliya TM (2022) In-silico computational investigations of antiviral Lignan derivatives as potent inhibitors of SARS CoV-2. ChemistrySelect 7(28):e202202069
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Jo S, Kim T, Iyer VG, Im W (2008) CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem 29(11):1859–1865
Vanommeslaeghe K, MacKerell AD Jr (2012) Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J Chem Inf Model 52(12):3144–3154
Baammi S, Daoud R, El Allali A (2023) Assessing the effect of a series of mutations on the dynamic behavior of phosphite dehydrogenase using molecular docking, molecular dynamics and quantum mechanics/molecular mechanics simulations. J Biomol Struct Dyn 41(9):4154–4166
Gumbart J, Khalili-Araghi F, Sotomayor M, Roux B (2012) Constant electric field simulations of the membrane potential illustrated with simple systems. Biochimica et Biophysica Acta (BBA) 1818(2):294–302
Bhandari S, Agrwal A, Kasana V, Tandon S, Boulaamane Y, Maurady A (2022) β-amino carbonyl derivatives: synthesis, molecular docking, ADMET, molecular dynamic and herbicidal studies. ChemistrySelect 7(48):e202201572
Boulaamane Y, Ahmad I, Patel H, Das N, Britel MR, Maurady A (2023) Structural exploration of selected C6 and C7-substituted coumarin isomers as selective MAO-B inhibitors. J Biomol Struct Dyn 41(6):2326–2340
Vegad UG, Gajjar ND, Nagar PR, Chauhan SP, Pandya DJ, Dhameliya TM (2023) In silico screening, ADMET analysis and MD simulations of phytochemicals of Onosma bracteata wall as SARS CoV-2 inhibitors. 3 Biotech 13(7):221
Dhameliya TM, Nagar PR, Gajjar ND (2022) Systematic virtual screening in search of SARS CoV-2 inhibitors against spike glycoprotein: pharmacophore screening, molecular docking, ADMET analysis and MD simulations. Mol Divers 26(5):2775–2792
Firth NC, Brown N, Blagg J (2012) Plane of best fit: a novel method to characterize the three-dimensionality of molecules. J Chem Inf Model 52(10):2516–2525
Sosnin S, Karlov D, Tetko IV, Fedorov MV (2018) Comparative study of multitask toxicity modeling on a broad chemical space. J Chem Inf Model 59(3):1062–1072
Medina-Franco JL, Martínez-Mayorga K, Giulianotti MA, Houghten RA, Pinilla C (2008) Visualization of the chemical space in drug discovery. Curr Comput Aided Drug Des 4(4):322–333
Shahapure KR, Nicholas C (2020) Cluster quality analysis using silhouette score. In: 2020 IEEE 7th international conference on data science and advanced analytics (DSAA), pp 747–748. IEEE
Rousseeuw PJ (1987) Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J Comput Appl Math 20:53–65
Rosén J, Gottfries J, Muresan S, Backlund A, Oprea TI (2009) Novel chemical space exploration via natural products. J Med Chem 52(7):1953–1962
Pollastri MP (2010) Overview on the rule of five. Curr Protoc Pharmacol 49(1):9–12
Zhang L, Tan J, Han D, Zhu H (2017) From machine learning to deep learning: progress in machine intelligence for rational drug discovery. Drug Discov Today 22(11):1680–1685
Yang J, Hu J, Zhang G, Qin L, Wen H, Tang Y (2021) Pharmacophore modeling and 3D-QSAR study for the design of novel α-synuclein aggregation inhibitors. J Mol Model 27(9):260
Vittorio S, Adornato I, Gitto R, Peña-Díaz S, Ventura S, De Luca L (2020) Rational design of small molecules able to inhibit α-synuclein amyloid aggregation for the treatment of Parkinson’s disease. J Enzyme Inhib Med Chem 35(1):1727–1735
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25(13):1605–1612
Vats S, Kondabala R, Saxena S (2022) Identification of alpha-Synuclein disaggregator from Camellia sp. insight of molecular docking and molecular dynamics simulations. ChemistrySelect 7(10):e202104131
Mohankumar T, Chandramohan V, Lalithamba HS, Jayaraj RL, Kumaradhas P, Sivanandam M, Elangovan N (2020) Design and molecular dynamic investigations of 7, 8-dihydroxyflavone derivatives as potential neuroprotective agents against alpha-synuclein. Sci Rep 10(1):599
Boulaamane Y, Kandpal P, Chandra A, Britel MR, Maurady A (2023) Chemical library design, QSAR modeling and molecular dynamics simulations of naturally occurring coumarins as dual inhibitors of MAO-B and AChE. J Biomol Struct Dyn 1–18
Kufareva I, Abagyan R (2012) Methods of protein structure comparison. Homol Model 231–257
Taidi L, Maurady A, Britel MR (2022) Molecular docking study and molecular dynamic simulation of human cyclooxygenase-2 (COX-2) with selected eutypoids. J Biomol Struct Dyn 40(3):1189–1204
Durham E, Dorr B, Woetzel N, Staritzbichler R, Meiler J (2009) Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J Mol Model 15:1093–1108
Haider S, Parkinson GN, Neidle S (2008) Molecular dynamics and principal components analysis of human telomeric quadruplex multimers. Biophys J 95(1):296–311
David CC, Jacobs DJ (2014) Principal component analysis: a method for determining the essential dynamics of proteins. Protein Dyn 193–226
Acknowledgements
Kailash Jangid gratefully acknowledge NSM for the access to ‘PARAM Seva Facility’ at IIT, Hyderabad, which is implemented by C-DAC and supported by the ministry of Electronics and Information Technology (MeitY) and Department of Science and Technology (DST), Government of India.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Author information
Authors and Affiliations
Contributions
YB suggested the idea, wrote the main manuscript text and performed the data analysis, KJ performed the MD simulations, MRB and AM supervised the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Boulaamane, Y., Jangid, K., Britel, M.R. et al. Probing the molecular mechanisms of α-synuclein inhibitors unveils promising natural candidates through machine-learning QSAR, pharmacophore modeling, and molecular dynamics simulations. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10691-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10691-x