Abstract
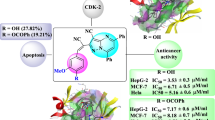
Gastric cancer is one of the malignant tumors of the gastrointestinal tract that, despite its decrease in recent years, is still the fourth most common cancer and the second leading cause of cancer-related death. Various strategies including chemotherapy are used to keep cancer cells from spreading and induce apoptotic death in them. Recent studies have shown that dihydropyrimidinones (DHPMs) are privileged structures in medicinal chemistry due to their pharmacological effects. A number of new 2-aminothiazolyl/benzothiazolyl derivatives of 3,4-DHPMs (3–8) were synthesized and structurally identified, and then their effects on the migration behavior of human AGS cells (gastric cancer cells) were investigated. Molecular docking and molecular dynamics (MD) simulations were applied to explore binding potential and realistic binding model of the assessed derivatives through identification of key amino acid residues within L5/α2/α3 allosteric site of kinesin 5 (Eg5) as a validated microtubule-dependent target for monastrol as a privileged DHPM derivative.
Graphical abstract













Similar content being viewed by others
Availability of data and material
All the data are available and can be offered on request.
Code availability
Not applicable.
References
Pecorino L (2016) Molecular biology of cancer: mechanisms, targets, and therapeutics. Oxford University Press, New York
Mons U, Gredner T, Behrens G, Stock C, Brenner H (2018) Cancers due to smoking and high alcohol consumption. Dtsch Arztebl Int 115(35–36):571–577
Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ (2009) Gastric cancer. Lancet 374(9688):477–490
Jiang F, Shen X (2019) Current prevalence status of gastric cancer and recent studies on the roles of circular RNAs and methods used to investigate circular RNAs. Cell Mol Biol Lett 24:53
Seyfried TN, Huysentruyt LC (2013) On the origin of cancer metastasis. Crit Rev Oncog 18(1–2):43–73
Kobayashi H, Enomoto A, Woods SL, Burt AD, Takahashi M, Worthley DL (2019) Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenter Hepatol 16(5):282–295
Alberts S, Cervantes A, Van de Velde C (2003) Gastric cancer: epidemiology, pathology and treatment. Ann Onc 14:ii31–ii36
Wang F, Li T, Zhang B, Li H, Wu Q, Yang L, Nie Y, Wu K, Shi Y, Fan D (2013) MicroRNA-19a/b regulates multidrug resistance in human gastric cancer cells by targeting PTEN. Biochem Biophys Res Com 434(3):688–694
Bidram Z, Sirous H, Khodarahmi GA, Hassanzadeh F, Dana N, Hariri AA, Rostami M (2020) Monastrol derivatives: in silico and in vitro cytotoxicity assessments. Res Pharm Sci 15(3):249–262
Sawin KE, LeGuellec K, Philippe M, Mitchison TJ (1992) Mitotic spindle organization by a plus-end-directed microtubule motor. Nature 359(6395):540–543
Pérez-Melero C (2014) KSP inhibitors as antimitotic agents. Curr Top Med Chem 14(20):2286–2311
Luo L, Carson JD, Dhanak D, Jackson JR, Huang PS, Lee Y, Sakowicz R, Copeland RA (2004) Mechanism of inhibition of human KSP by monastrol: insights from kinetic analysis and the effect of ionic strength on KSP inhibition. Biochemistry 43(48):15258–15266
González-Hernández E, Aparicio R, Garayoa M, Montero MJ, Sevilla MÁ, Pérez-Melero C (2019) Dihydropyrimidine-2-thiones as Eg5 inhibitors and L-type calcium channel blockers: potential antitumour dual agents. MedChemComm 10(9):1589–1598
Nakai R, Iida S, Takahashi T, Tsujita T, Okamoto S, Takada C, Akasaka K, Ichikawa S, Ishida H, Kusaka H, Akinaga S, Murakata C, Honda S, Nitta M, Saya H, Yamashita Y (2009) K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res 69(9):3901–3909
Khathi SP, Chandrasekaran B, Karunanidhi S, Tham CL, Kozielski F, Sayyad N, Karpoormath R (2018) Design and synthesis of novel thiadiazole-thiazolone hybrids as potential inhibitors of the human mitotic kinesin Eg5. Bioorg Med Chem Lett 28(17):2930–2938
Van Meerloo J, Kaspers GJ, Cloos J (2011) Cell sensitivity assays: the MTT assay. In: Cancer cell culture, Humana Press
Sun XD, Shi XJ, Sun XO, Luo YG, Wu XJ, Yao CF, Yu HY, Li DW, Liu M, Zhou J (2011) Dimethylenastron suppresses human pancreatic cancer cell migration and invasion in vitro via allosteric inhibition of mitotic kinesin Eg5. Acta Pharm Sin 32(12):1543
Wang X, Decker CC, Zechner L, Krstin S, Wink M (2019) In vitro wound healing of tumor cells: inhibition of cell migration by selected cytotoxic alkaloids. BMC Pharm Toxic 20(1):1–2
Morris G, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Sanner M (1999) Python: a programming language for software integration and development. J Mol Graph Model 17(1):57–61
Razzaghi-Asl N, Ebadi A, Shahabipour S, Gholamin D (2020) Identification of a potential SARS-CoV2 inhibitor via molecular dynamics simulations and amino acid decomposition analysis. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2020.1797536
Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M (2015) PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res 43(W1):W443–W447
Kaan HYK, Ulaganathan V, Rath O, Prokopcova H, Dallinger D, Kappe CO, Kozielski F (2010) Structural basis for inhibition of Eg5 by dihydropyrimidines: stereoselectivity of antimitotic inhibitors enastron, dimethylenastron and fluorastrol. J Med Chem 53:5676–5683
Spoel VDD, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Schuttelkopf AW, Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60(8):1355–1356
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys 52(12):7182–7190
Hess B, Bekker H, Berendsen HJC, Fraaije J (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: An N log (N) method for Ewald sums in large systems. J Chem Phys 98(12):10089–10092
Razzaghi-Asl N, Mirzayi S, Mahnam K, Sepehri S (2018) Identification of COX-2 inhibitors via structure-based virtual screening and molecular dynamics simulation. J Mol Graph Model 83:138–152
Bohlooli S, Nejatkhah N, Sepehri S, Doostkamel D, Razzaghi-Asl N (2020) Synthesis and cytotoxicity evaluation of novel cyclic/noncyclic N-heteroaryl enamino amides vs human cancer cell lines. Res Pharm Sci 15(6):563–570
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717
Kapitein LC, Peterman EJG, Kwok BH, Kim JH, Kapoor TM, Schmidt CF (2005) The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature 435(7038):114–118
Sakowicz R, Finer JT, Beraud C, Crompton A, Lewis E, Fritsch A, Lee Y, Mak J, Moody R, Turincio R, Chabala JC, Gonzales P, Roth S, Weitman S, Wood KW (2004) Antitumor activity of a kinesin inhibitor. Cancer Res 64(9):3276–3280
Imai T, Oue N, Nishioka M, Mukai S, Oshima T, Sakamoto N, Sentani K, Matsusaki K, Yoshida K, Yasui W (2017) Overexpression of KIF11 in gastric cancer with intestinal mucin phenotype. Pathobiology 84:16–24
Lu M, Zhu H, Wang X, Zhang D, Xiong L, Xu L, You Y (2016) The prognostic role of Eg5 expression in laryngeal squamous cell carcinoma. Pathology 48(3):214–218
Myers SM, Collins I (2016) Recent findings and future directions for interpolar mitotic kinesin inhibitors in cancer therapy. Future Med Chem 8(4):463–489
Garcia-Saez I, DeBonis S, Lopez R, Trucco F, Rousseau B, Thuery P, Kozielski F (2007) Structure of human Eg5 in complex with a new monastrol-based inhibitor bound in the R configuration. J Biol Chem 282:9740–9747
Durrant JD, McCammon JA (2011) Molecular dynamics simulations and drug discovery. BMC Biol 9:71–79
Acknowledgements
The research leading to offered results received funding from Ardabil University of Medical Sciences (ARUMS) under Grant No. IR.ARUMS.REC.1398.259.
Funding
The research leading to offered results received funding from Ardabil University of Medical Sciences (ARUMS) under Grant No. IR.ARUMS.REC.1397.137.
Author information
Authors and Affiliations
Contributions
All authors contributed equally in this work (concept, design, definition of intellectual content, literature search, experimental studies, data acquisition, data analysis, statistical analysis, manuscript preparation, manuscript editing and manuscript review).
Corresponding author
Ethics declarations
Conflict of interest
Authors declare to have no conflicts of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sagha, M., Mousaei, F., Salahi, M. et al. Synthesis of new 2-aminothiazolyl/benzothiazolyl-based 3,4-dihydropyrimidinones and evaluation of their effects on adenocarcinoma gastric cell migration. Mol Divers 26, 1039–1051 (2022). https://doi.org/10.1007/s11030-021-10229-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-021-10229-z