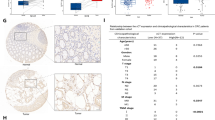
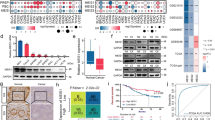
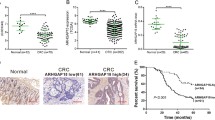
Abstract
Colorectal cancer (CRC) is the third most commonly diagnosed cancer worldwide. Chemotherapy is the mainstay of treatment for patients with CRC in II-IV stages. Resistance to chemotherapy occurs commonly, which results in treatment failure. Therefore, the identification of novel functional biomarkers is essential for recognizing high-risk patients, predicting recurrence, and developing new therapeutic strategies. Herein, we assessed the roles of KIAA1549 in promoting tumor development and chemoresistance in colorectal cancer. As a result, we found that KIAA1549 expression is up-regulation in CRC. Public databases revealed a progressive up-regulation of KIAA1549 expression from adenomas to carcinomas. Functional characterization uncovered that KIAA1549 promotes tumor malignant phenotypes and boosts the chemoresistance of CRC cells in an ERCC2-dependent manner. Inhibition of KIAA1549 and ERCC2 effectively enhanced the sensitivity to chemotherapeutic drugs oxaliplatin and 5-fluorouracil. Our findings suggest that endogenous KIAA1549 might function as a tumor development-promoting role and trigger chemoresistance in colorectal cancer partly by upregulating DNA repair protein ERCC2. Hence, KIAA1549 could be an effective therapeutic target for CRC and inhibition of KIAA1549 combined with chemotherapy might be a potential therapeutic strategy in the future.






Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Sinicrope FA (2022) Increasing incidence of early-onset colorectal cancer. N Engl J Med 386:1547–1558. https://doi.org/10.1056/NEJMra2200869
Pastushok L, Fu Y, Lin L et al (2019) A novel cell-penetrating antibody Fragment inhibits the DNA repair protein RAD51. Sci Rep. https://doi.org/10.1038/s41598-019-47600-y
Gustavsson B, Carlsson G, Machover D et al (2015) A review of the evolution of systemic chemotherapy in the management of colorectal cancer. Clin Colorectal Cancer 14:1–10. https://doi.org/10.1016/j.clcc.2014.11.002
Mellor HR, Callaghan R (2008) Resistance to chemotherapy in cancer: a complex and integrated cellular response. Pharmacology 81:275–300. https://doi.org/10.1159/000115967
Biagi JJ, Raphael MJ, Mackillop WJ et al (2011) Association between time to initiation of adjuvant chemotherapy and survival in colorectal cancer: a systematic review and meta-analysis. JAMA 305:2335–2342. https://doi.org/10.1001/jama.2011.749
Jeuken JW, Wesseling P (2010) MAPK pathway activation through BRAF gene fusion in pilocytic astrocytomas; a novel oncogenic fusion gene with diagnostic, prognostic, and therapeutic potential. J Pathol 222:324–328. https://doi.org/10.1002/path.2780
Jones DTW, Kocialkowski S, Liu L et al (2008) Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res 68:8673–8677. https://doi.org/10.1158/0008-5472.CAN-08-2097
Jain P, Silva A, Han HJ et al (2017) Overcoming resistance to single-agent therapy for oncogenic BRAF gene fusions via combinatorial targeting of MAPK and PI3K/mTOR signaling pathways. Oncotarget. https://doi.org/10.18632/oncotarget.20949
Chen R, Keoni C, Waker CA et al (2019) KIAA1549-BRAF expression establishes a permissive tumor microenvironment through NFκB-mediated CCL2 production. Neoplasia 21:52–60. https://doi.org/10.1016/j.neo.2018.11.007
Kumar A, Pathak P, Purkait S et al (2015) Oncogenic KIAA1549-BRAF fusion with activation of the MAPK/ERK pathway in pediatric oligodendrogliomas. Cancer Genet 208:91–95. https://doi.org/10.1016/j.cancergen.2015.01.009
Kaul A, Chen Y-H, Emnett RJ et al (2012) Pediatric glioma-associated KIAA1549:BRAF expression regulates neuroglial cell growth in a cell type-specific and mTOR-dependent manner. Genes Dev 26:2561–2566. https://doi.org/10.1101/gad.200907.112
Karajannis MA, Legault G, Fisher MJ et al (2014) Phase II study of sorafenib in children with recurrent or progressive low-grade astrocytomas. Neuro-Oncol 16:1408–1416. https://doi.org/10.1093/neuonc/nou059
Subbiah V, Westin SN, Wang K et al (2014) Targeted therapy by combined inhibition of the RAF and mTOR kinases in malignant spindle cell neoplasm harboring the KIAA1549-BRAF fusion protein. J Hematol OncolJ Hematol Oncol 7:8. https://doi.org/10.1186/1756-8722-7-8
Li N, Zhan X (2019) Identification of clinical trait–related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA J 10:273–290. https://doi.org/10.1007/s13167-019-00175-0
Lin M, Fang Y, Li Z et al (2021) S100P contributes to promoter demethylation and transcriptional activation of SLC2A5 to promote metastasis in colorectal cancer. Br J Cancer 125:734–747. https://doi.org/10.1038/s41416-021-01306-z
Antonelli M, Badiali M, Moi L et al (2015) KIAA1549:BRAF fusion gene in pediatric brain tumors of various histogenesis. Pediatr Blood Cancer 62:724–727. https://doi.org/10.1002/pbc.25272
Nagase T (2000) Prediction of the coding sequences of unidentified Human GENES. XVIII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res 7:271–281. https://doi.org/10.1093/dnares/7.4.271
Gupta S, Provenzale D, Llor X et al (2019) NCCN guidelines insights: genetic/Familial High-Risk assessment: colorectal, version 2.2019. J Natl Compr Cancer Netw JNCCN 17:1032–1041. https://doi.org/10.6004/jnccn.2019.0044
Kopetz S, Hoff PM, Morris JS et al (2010) Phase II trial of infusional fluorouracil, irinotecan, and bevacizumab for metastatic colorectal cancer: efficacy and circulating angiogenic biomarkers associated with therapeutic resistance. J Clin Oncol Off J Am Soc Clin Oncol 28:453–459. https://doi.org/10.1200/JCO.2009.24.8252
Graf N, Ang WH, Zhu G et al (2011) Role of endonucleases XPF and XPG in nucleotide excision repair of platinated DNA and cisplatin/oxaliplatin cytotoxicity. ChemBioChem 12:1115–1123. https://doi.org/10.1002/cbic.201000724
Yuan X, Zhang W, He Y et al (2020) Proteomic analysis of cisplatin- and oxaliplatin-induced phosphorylation in proteins bound to Pt-DNA adducts. Met Integr Biometal Sci 12:1834–1840. https://doi.org/10.1039/d0mt00194e
Metzger R, Leichman CG, Danenberg KD et al (1998) ERCC1 mRNA levels complement thymidylate synthase mRNA levels in predicting response and survival for gastric cancer patients receiving combination cisplatin and fluorouracil chemotherapy. J Clin Oncol 16:309–316. https://doi.org/10.1200/JCO.1998.16.1.309
Welsh C, Day R, McGurk C et al (2004) Reduced levels of XPA, ERCC1 and XPF DNA repair proteins in testis tumor cell lines. Int J Cancer 110:352–361. https://doi.org/10.1002/ijc.20134
Dabholkar M, Vionnet J, Bostick-Bruton F et al (1994) Messenger RNA levels of XPAC and ERCC1 in ovarian cancer tissue correlate with response to platinum-based chemotherapy. J Clin Invest 94:703–708. https://doi.org/10.1172/JCI117388
Berndt SI, Platz EA, Fallin MD et al (2006) Genetic variation in the nucleotide excision repair pathway and colorectal cancer risk. Cancer Epidemiol Prev Biomark 15:2263–2269. https://doi.org/10.1158/1055-9965.EPI-06-0449
Gurubhagavatula S, Liu G, Park S et al (2004) XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non-small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol 22:2594–2601. https://doi.org/10.1200/JCO.2004.08.067
Joo J, Yoon K-A, Hayashi T et al (2016) Nucleotide excision repair gene ERCC2 and ERCC5 variants increase risk of uterine cervical cancer. Cancer Res Treat 48:708–714. https://doi.org/10.4143/crt.2015.098
Lee M-S, Liu C, Su L, Christiani DC (2015) Polymorphisms in ERCC1 and ERCC2/XPD genes and carcinogen DNA adducts in human lung. Lung Cancer 89:8–12. https://doi.org/10.1016/j.lungcan.2015.05.001
Boldrin E, Malacrida S, Rumiato E et al (2019) Association between ERCC1 rs3212986 and ERCC2/XPD rs1799793 and OS in patients with advanced esophageal cancer. Front Oncol. https://doi.org/10.3389/fonc.2019.00085
Li Q, Damish AW, Frazier Z et al (2019) ERCC2 helicase domain mutations confer nucleotide excision repair deficiency and drive cisplatin sensitivity in muscle-invasive bladder cancer. Clin Cancer Res 25:977–988. https://doi.org/10.1158/1078-0432.CCR-18-1001
Damia G, Guidi G, D’Incalci M (1998) Expression of genes involved in nucleotide excision repair and sensitivity to cisplatin and melphalan in human cancer cell lines. Eur J Cancer 34:1783–1788. https://doi.org/10.1016/S0959-8049(98)00190-7
Zhao H, Yu X, Ding Y et al (2016) MiR-770–5p inhibits cisplatin chemoresistance in human ovarian cancer by targeting ERCC2. Oncotarget 7:53254–53268. https://doi.org/10.18632/oncotarget.10736
O’Grady S, Finn SP, Cuffe S, Richard DJ, O’Byrne KJ, Barr MP (2014) The role of DNA repair pathways in cisplatin resistant lung cancer. Cancer Treat Rev 40(10):1161–70. https://doi.org/10.1016/j.ctrv.2014.10.003
Weaver DA, Crawford EL, Warner KA, et al (2005) ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer 4:18. https://doi.org/10.1186/1476-4598-4-18
Acknowledgements
We thank all laboratory members of the Department of Pathology of Southern Medical University for their support and comments. The schematic graph is drawn by FigDraw.
Funding
This study was supported by National Natural Science Foundation of China (82273564, 82073342, 82103595); Science and Technology Projects in Guangzhou, China (202206010045, 202201010911); Guangdong Provincial Regional Joint Fund-Youth Fund Project (2020A1515110006); The Foundation of President of Nanfang Hospital (2020C039, 2020C033, 2020C006, 2020B012, 2019B009).
Author information
Authors and Affiliations
Contributions
FY, YX, ML: Conceptualization, Methodology, Software, Writing-Original draft preparation. YL, YF: Writing—Reviewing and Editing. KC: Supervision. YZ, YD: Project administration. All authors discussed the results and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Ethics Committee of Nanfang Hospital of Southern Medical University (NEFC-2022-367).
Consent to publish
All authors read and approved the fnal manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ye, F., Xie, Y., Lin, M. et al. KIAA1549 promotes the development and chemoresistance of colorectal cancer by upregulating ERCC2. Mol Cell Biochem 479, 629–642 (2024). https://doi.org/10.1007/s11010-023-04751-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-023-04751-x