Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been prevalent in the humans since 2019 and has given rise to a pandemic situation. With the discovery and ongoing use of drugs and vaccines against SARS-CoV-2, there is still no surety of its complete suppression of this disease or if there is a need for additional booster doses. There is an urgent need for alternative treatment strategies against COVID-19. Peptides and peptidomimetics have several advantages as therapeutic agents because of their target selectivity, better interactions, and lower toxicity. Minor structural alterations to peptides can help prevent their fast metabolism and provide long-action. This comprehensive review provides an overview of different peptide-based vaccines and therapeutics against SARS-CoV-2. It discusses the design and mechanism of action of the peptide-based vaccines, peptide immunomodulators, anti-inflammatory agents, and peptides as entry inhibitors of SARS-CoV-2. Moreover, the mechanism of action, sequences and current clinical trial studies are also summarized. The review also discusses the future aspects of peptide-based vaccines and therapeutics for COVID-19.
Graphical Abstract

Similar content being viewed by others
Introduction
The rise of novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has grasped the entire world with restless and questionable situation because of its high pace of human-to-human transmission of infection (Li et al. 2020). COVID-19 is caused due to the association between SARS-CoV-2 and the host surface receptor protein, human angiotensin-convertase enzyme 2 (hACE2) (Shang et al. 2020). Therefore, hACE2 can be considered the primary cell passage site for SARS-CoV-2 (Li et al. 2020). SARS-CoV-2 is known to enter through two essential passageways, nose, and mouth, and reaches the lungs, cerebrum, and digestive system, where there is plethora of hACE2. As the virus enters the host cell, it affects several organ system functions, which leads to severe illness. The higher pace of ailment and mortality hazard is profoundly connected with the senior citizens, patients with debilitating health conditions, and people already affected with diseases like diabetes, malignant growth, lung illness, hypertension, etc (Li et al. 2020). As COVID-19 has become a pandemic, researchers worldwide are in a hassle to comprehend the COVID-19 adversary and find a reasonable treatment strategy to beat this disease (https://www.oecd.org/coronavirus/policy-responses/testing-for-covid-19-a-way-to-lift-confinement-restrictions-89756248/).
To date, no clinically endorsed treatments can forestall COVID-19 completely. Subsequently, the underlying step is to look for the prospective targets from existing drugs such as remdesivir, lopinavir, chloroquine, hydroxychloroquine, etc., and so forth to treat SARS-CoV-2, yet their viability is difficult (Gordon et al. 2020). The symptoms of COVID-19 vary from person to person. People have reported various symptoms ranging from mild cold, cough, fever to being asymptomatic. The worst symptoms in some patients are cytokine storm and acute respiratory distress syndrome. These symptoms increase the mortality to morbidity ratio (Yi et al. 2020). Various strategies are employed in the drug discovery process against COVID-19. In this regard, peptide therapeutics promises a potential against COVID-19. Peptides are parts of the proteins and are ideal for their simple synthesis as far as time and cost are considered (Tizzano et al. 2005). The gateway of advancement for peptide-based therapeutics was broadly opened by modifying the peptides within the parameters such as sequence length, side-chain reactivity, and other unnatural parts. The benefits of peptides as medications are their specificity towards the target site and low metabolic toxicity because of the restricted opportunities for aggregation in the body (Bruno et al. 2013; Muttenthaler et al. 2021). We discuss various strategies that are being explored for tackling COVID-19 viral infection, such as peptide vaccines, peptide immune modulators, anti-inflammatory peptides, and entry inhibiting peptides. Currently, there is no precise treatment for COVID-19. Thus, it is vital to identify prospective targets to build and repurpose useful drugs. Peptide-based vaccines, immune modulators, anti-inflammatory, and entry inhibiting strategies are recently researched (Ahmed et al. 2019; Kalita et al. 2020; Kurpe et al. 2020; Mahendran et al. 2020; Nyanguile 2019).
This comprehensive review provides an overview of the design and mechanism of the strategies mentioned above. We also collectively enlist the different repurposing or designing studies in clinical trials and several pharmaceutical companies working on the same.
Peptide-Based Vaccines for COVID-19
Peptide-based vaccines (PV) are synthetic vaccines (Nevagi et al. 2018). They are synthesized in vitro using known immunogenic amino acids (~ 20 to 30 amino acids), which resemble antigenic sites and can trigger immune responses. PV are expected to be a safer alternative to traditional vaccines. Large-scale production of these PV would be easier at low cost and high reproducibility rate due to their chemical synthesis (Nevagi et al. 2018). PV are water-soluble and more stable compared to conventional vaccines (CV). The body enzymes can quickly degrade PV before eliciting an immune stimulation. Thus, they are known to be weak immunogens. Like the CV, PV requires adjuvants to increase the vaccine efficacy (Purcell et al. 2007). PV are generally made of laboratory synthesized B or T cell epitopes. T cells identify the peptide sequences complexed with the antigen presentation molecules, major histocompatibility complex (MHC) I or II classes on the antigen-presenting cell surface. CD4 helper T cells work with B cells to produce antibodies against the infection and help the CD8 cytotoxic cells to kill the infected cells (Natale et al. 2020). Epitope mapping involves identifying the binding site of the antibodies on the antigen using in silico computational models and then optimizing these epitopes to enhance their functional properties to elicit a better immune response. Once these epitopes are identified and optimized, they can be loaded into various delivery systems (such as nanoparticles, dendrimers, liposomes) to prevent their degradation (Natale et al. 2020). PV has been under investigation for several decades. A quick search in clinicaltrials.gov with keywords 'peptide-based vaccines' revealed 27 studies at different stages of their clinical trials, and of which 14 are completed (as of October 2021) (Bar-Natan 2021; Bionor Immuno 2017; CENTOGENE GmbH Rostock 2021; Centre Hospitalier Universitaire de Besancon 2021; Cliniques universitaires Saint-Luc- Université Catholique de Louvain 2020; Craig et al. 2021; Dietrich 2016; Disis 2017; George Peoples and MD, FACS, 2020; Herbert Irving Comprehensive Cancer Center 2013; Immatics Biotechnologies GmbH 2014; Jr 2016a, b, c, 2020; Mayo Clinic 2018; OSE Immunotherapeutics 2021; Southwest Oncology Group 2015; United Neuroscience Ltd 2020; University of Southern California 2014; University of Washington 2017, 2019, 2020). In this article, we are reviewing several PV strategies against the SARS-CoV-2.
One of the several approaches in designing a PV for COVID-19 was using the cytotoxic T lymphocyte MHC class I epitope. In a recent study on the Ebola Virus (EBOV) infection, a nine amino-acid peptide, NP44-52, was immunogenic in vivo. The most targeted site in the survivors (CTL) was the EBOV-NP43-53 epitope. Thus, a smaller fragment of the peptide was found to be immunogenic (Herst et al. 2020). Based on these experiments, it was hypothesized that similar CTL could be beneficial for coronavirus infections (Channappanavar et al. 2014). Thus, several immunoproteomic studies were conducted to identify potential immunogenic peptides on SARS-CoV-2 and SARS-CoV-2 nucleocapsid proteins. The nucleocapsid phosphoprotein generated a lot of antibodies in the SARS-infected patients (Herst et al. 2020). Khan et al. reported a systematic study on identifying MHC I/MHC II binding epitopes, and their screening tool provided three MHC I and five MHC II epitopes (Alam et al. 2021).
Similarly, several other multi-epitope approaches were also applied for developing vaccine candidates for SARS-CoV-2. Prachar et al. reported 174 epitopes with high prediction binding scores (Prachar et al. 2020). The epitopes were validated to bind to 11 HLA allotypes MHC I/II. These were tested against the SARS-CoV-E protein as the target antigen. The multi-epitope T-cell-based peptide vaccine was found to be a promising vaccine candidate (Abdelmageed et al. 2020).
Apart from the in-silico modeling, several in vivo proteomic investigations were conducted. The antibody response to the rapid-binding domain (RBD) and the S proteins was assessed in vivo. The study was conducted on blood collected from COVID-19 infected convalescent individuals (149 individuals). The plasmas (Day 39 of infection) were tested by using ELISA for their ability to bind to SARS-CoV-2 RBD and S protein sites. The nature of these antibodies was found out by isolating the B cells. Several monoclonal antibodies were expressed in cells, and their neutralizing activity was quantified by ELISA. They observed multiple neutralizing epitopes on the RBD identified and neutralized by the pseudo virus. Thus, the analyzed cohort (6 individuals) did not contain high neutralizing antibodies but had RBD-specific antibodies with antiviral activity. These observations suggest that a vaccine design that elicits these RBD-specific antibodies should be effective (Robbiani et al. 2020). Similarly, in another study by Poh et al., two immunodominant linear B-cell epitopes (S14 and S21) against the SARS-CoV-2 S glycoprotein were reported (Poh et al. 2020).
Along with the peptide-based vaccines, several neutralizing monoclonal antibodies against SARS-CoV-2 are also being developed (Taylor et al. 2021). The monoclonal antibodies are a novel class of antiviral therapeutics. They can bind to and neutralize the virus. They are recombinant proteins derived from humanized mice or convalescent patients (B cells). Monoclonal antibody therapy is a type of passive immunotherapy. There are several studies related to COVID-19 peptide-based vaccines currently in the clinical trials (Table 1). Also, many companies are developing peptide-based vaccines against SARS-CoV-2 (Table 2). Here, we have collectively summarized all the peptide vaccine studies.
Peptide Immune Modulators
Immune modulators are agents that work on the checkpoints in the pathways of the immune system. They modify, increase, or reduce the immune response, which can help reduce chronic symptoms. Increased levels of anti-inflammatory cytokines (primarily IL-4 and IL-10) and Treg cell populations restrict immune cell generation and activation, leading to a long-term reduction in immune cell numbers and functions. As a result, patients become vulnerable to various secondary opportunistic infections, aggravating disease development. Immune enhancement therapy given at the right time can help prevent such instances (Zhang et al. 2020). Here, we discuss the peptide-based immune enhancement and modulation therapies used in COVID-19. The immune suppression, anti-inflammatory class of peptide-based therapies is discussed under a separate heading further.
Immunological enhancement therapy administered on time can correct immune deficiencies and increase immunity. Thymosin and human gamma globulin are some of the treatments available. Thymosin is a polypeptide produced by the thymus endogenously in the human body. It has been used to treat viral infections as an immune system modifier for many years. It is reported to restore the immune system exhaustion and potentiates differentiation of immune cells. Its activity in COVID-19 is still not clear (Liu et al. 2020). When an immunological response is required, lymphocytes and plasma cells of the immune system create human gamma globulin. Immunoglobulins, often known as antibodies, are globulins that aid in immunological responses and immunity, and almost all forms of human gamma globulin are immunoglobulins (Human Gamma Globulin 2021).
RTD-1 is an antiviral immunomodulator that activates protective immunity (Mahendran et al. 2020). It leads to a blunted pro-inflammatory cytokine response causing decreased overexpression of cytokine levels of IL-6, keratinocyte chemo-attractant, and GM-CSF in lung cell homogenates and helps in eliminating SARS-CoV. The term associated with overexpression of inflammatory response of the immune system is “cytokine storm” (Ruscitti et al. 2020). CIGB-258 is an immunomodulatory peptide for the treatment of COVID-19 (Dominguez-Horta and Del 2020). Several studies on COVID-19 peptide-based immune modulators in clinical trials (Table 3).
Anti-inflammatory Peptides/Immune Suppression
Anti-inflammatory peptides [such as interleukin (IL) IL-4, IL-10, IL-13, IL-35, transforming growth factor-β (TGF-β), TGF-p1] are a class of small intracellular peptides (5–140 kD) that are endogenously released by the body in response to control the immune response. Anti-inflammatory peptides block the production of inflammatory mediators (TNF, IL-1, IL-10, and TGF- β) and inhibit the synthesis and release of pro-inflammatory cytokines (IL-6) and exert a protective effect on the body in most infectious diseases (Manna et al. 2018). Many peptides of herbal, mammalian, and bacterial origin have been reported to possess anti-inflammatory activity. Synthetic anti-inflammatory peptides are also reported (Dadar et al. 2019).
Inflammation is a mechanism used by the human body to defend the infection. Overexpression of inflammatory mechanisms causes increased production of inflammatory molecules, such as oxidants, pro-inflammatory cytokines, chemokines, matrix metalloproteinases, and transcription factors in the damaged tissues. This is the starting point of many chronic diseases, several endogenous peptides released in the body during the inflammatory process exhibit anti-inflammatory activity. Endogenous and synthetic peptides exhibiting anti-inflammatory activity are already reported and can be explored further to treat COVID-19 infection-associated overexpression of inflammatory mechanisms. SARS-CoV-2 infection can lead to varied responses, such as being asymptomatic to facing chronic severe symptoms. Considering individuals' genetic composition and immune system response, the symptoms can further progress to interstitial pneumonia or acute respiratory distress syndrome. These clinical symptoms are seen in patients with old age or comorbidities. This subgroup of patients is notable for having very high levels of serum ferritin and D-dimer levels, hepatic dysfunction, thrombotic tendency, and disseminated intravascular coagulation (DIC), implicating occurrence of macrophage activation syndrome (MAS), also known as secondary hemophagocytic lymph histiocytosis (sHLH), cytokine storm, multiple organ failure and eventually death (Soy et al. 2020; Zhou et al. 2020). Antiviral drugs alone will not be sufficient for the treatment of COVID-19. Anti-inflammatory agents are explored for COVID-19 along with antiviral therapy. They have profoundly helped provide symptomatic relief. The anti-inflammatory peptides market has seen a significant surge during the 2020 global pandemic (Global Anti-Inflammatory Peptides Market Size 2021).
Anti-inflammatory peptides decrease synthesis, release, and inhibit the pro-inflammatory cytokines and inflammatory mediators, respectively, thereby providing great relief to the condition caused by a cytokine storm. The synthetic anti-inflammatory peptides (peptidomimetics) are designed based on the endogenous peptides; they have similarities with their endogenous counterparts (Dadar et al. 2019; Du et al. 2009a; Sala et al. 2018).
In COVID-19 infection, the significant observed clinical findings are increased total neutrophils (N) and lymphocytes (L), and N/L ratio increases, indicating hyperactivity and high infection. Besides this, the peripheral N and L count decreases, causing atrophy in the secondary organs like the spleen and lymph nodes, as reported from the autopsy of the COVID-19 infected patients. The number of monocytes and macrophages is also increased. On the other hand, this justifies the increase in pro-inflammatory cytokines such as IL-6, IL-1, tumor necrosis factor (TNF) α, and IL-8. Increased inflammatory markers lead to a condition of higher blood viscosity, mechanical ventilation, and vascular interventions, which progresses to multiple organ failure and death (Bruzzese and Lazzarino 2020; Mahmudpour et al. 2020; Manna et al. 2018; Roshanravan et al. 2020; Ruscitti et al. 2020; Zhou et al. 2020).
The virus binds to cellular receptors in pancreatic cells, causing functional abnormalities in infected pancreatic beta cells, leading to onset diabetes mellitus symptoms. Hence, GLP-1 is explored for having anti-inflammatory effects in COVID-19. It decreases the CRP levels that are increased in the infected condition and is reported to reduce complications of diabetes mellitus associated with COVID-19 (Katsiki and Ferrannini 2020; Lee et al. 2021; Sazgarnejad et al. 2021).
Interestingly according to a study report on COVID-19 biomarkers, CRP, IL-6, and dimers are the biomarkers explored for low glycemic index. Also, a low glycemic index is reported in elderly COVID-19 patients. Anti-diabetic drugs (peptides/peptidomimetics) include liraglutide, linaglutide, dulaglutide, semaglutide, lixientide, and exenatide, are also said to decrease these biomarkers. These peptidomimetics can be potential treatment options for COVID-19 (Katsiki and Ferrannini 2020).
TAT CARMIL1 is a drug made up of two naturally occurring peptides that, when combined, work to enter a cell membrane, and decrease an acute inflammatory response. The peptide may mitigate some of the harshest effects of acute inflammatory responses (https://www.genengnews.com/news/peptide-drug-discovery-could-lead-to-a-powerful-anti-inflammatory/). There are several studies on COVID-19 peptide-based anti-inflammatory agents in clinical trials (Table 4).
Entry Inhibiting Peptides/Inhibitory Peptides
There are currently no specific treatments for SARS-CoV-2 (Pouwels et al. 2021). In this concern, entry inhibition is the novel way to combat the coronavirus (Agrawal et al. 2020). Peptide inhibitors have considerable potential for treating newly developing viral infections due to their safety, effectiveness, and selectivity (Bruno et al. 2013). They are appealing alternatives to small molecules because peptides may be more efficient and specific than small molecule drugs and thus could be tolerated better. Antiviral peptides can be rationally developed and improved based on the known structures of viral proteins and their biological targets (Eggink et al. 2019). The peptides that arise may be particular for their targets and viral infections or have broad antiviral efficacy (Blank et al. 2016; Vagner et al. 2008). The current state of peptides that block SARS-CoV-2 entrance is summarized here and has described the methods for developing peptides that target the ACE2 receptor or the viral Spike protein and the activating proteases furin transmembrane serine protease 2 (TMPRSS2), or cathepsin L.
Entry of SARS-CoV-2
Seven coronaviruses (CoVs) have propagated and caused disease in humans after crossing species barriers from their bat reservoir via several intermediary hosts (Gorbalenya et al. 2020). The coronaviruses HCoV-229E, HCoV-NL63, HCoV-OC43 and HCoV-HKU1 have all been linked to endemic infections of the upper respiratory tract and result in common colds. In this fashion, in recent years, SARS-CoV-1, MERS-CoV (Middle East respiratory disease CoV), and, most recently, SARS-CoV-2 have all been recognised (Shang et al. 2020).
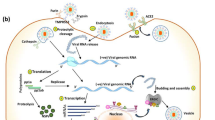
SARS-CoV-2 enters its target cells in several stages (Fig. 1). The S protein of SARS-S CoV-2 interacts with the SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2). Cellular proteases must activate the S protein to insert the viral fusion peptide into the host membrane, a process known as anchoring. The HR1 and HR2 portions of the trimeric viral transmembrane protein then interact to form a six-helix bundle that pulls the viral and cellular membranes together and causes fusion, allowing the viral genome to be released into the cytoplasm (Du et al. 2009b; Hoffmann et al. 2020a; Braun and Sauter 2019; Gur et al. 2020).
Mechanism of Entry Inhibition by Peptides
Peptides Preventing ACE2 Binding of the SARS-CoV-2 Spike Protein
The first step in the entry of SARS-CoV-2 is binding the viral S protein to its cellular receptor ACE2. Most of the peptides designed are based on the interactions of RDB binding to the ACE receptor (Hofmann et al. 2005). From the interpretations of Alanine scanning, it is observed that the residues ranging from 22 to 57 of ACE are responsible for its interaction with spike protein. Based on that, Hans and co-workers developed a set of six peptides from this region to bind them with the SARS-CoV-1 S1 RBD and block SARS-CoV-1 pseudo particle binding. Of them, P4, P5, and P6 have shown the IC50 values in the micromolar range, in which P6 was found to show higher potency up to 0.1 μM (Han et al. 2006).
Cao and co-workers developed peptides to inhibit SARS-CoV-2 using i) ACE2 as a scaffold and ii) de-novo sequencing based on RBD-binding interaction motifs. They have found the two critical candidates, AHB1 and AHB2, which inhibited SARS-CoV-2 with IC50 values of 35 and 16 nm, respectively. From the de-novo sequencing, they have identified LCB1 and LCB3, which bind the RBD with lower dissociation constants and inhibited SARS-CoV-2 in the picomolar range (Cao et al. 2020).
Peptides Targeting ACE2
ACE2 is a membrane-associated aminopeptidase found in the heart, blood vessels, lung, kidney, intestine, testis, and brain. SARS-COV-1 and SARS-COV-2 were found to enter using ACE2 as a receptor (Hamming et al. 2004; Verdecchia et al. 2020). With the help of predicted hydrophilicity, surface probability, and chain flexibility, Ho and co-workers developed a set of peptides derived from the S protein of SARS-CoV-1 such as SP-4 (192–203), SP-8(483–494), and SP-10(668–679) which are found to abrogate the interaction between a recombinant S protein and ACE2. The SP-10 has inhibited SARS-CoV-1 pseudo particle entry in Vero cells in the nanomolar range (Ho et al. 2006).
Antiviral Peptides Targeting Furin
Furin is a proprotein convertase widely found in eukaryotic tissues and physiologically cleaves precursors of multiple proteins, including growth factors, hormones, cell surface receptors, etc (Braun and Sauter 2019). The SARS-CoV-2 is different from SARS-CoV-1 in activating SARS-CoV-2 S protein by furin through cleavage at a polybasic cleavage site into its S1-S2 subunits. A ketone-based peptide, Decanoyl-RVKR chloromethyl ketone (dec-RVKR-cmk), is a well-established peptidomimetic furin inhibitor, which inhibits furin by a chemical reaction resulting in hemiketal and found to show the irreversible inhibition of its enzymatic activity (Becker et al. 2010; Bergeron et al. 2005; Sugrue et al. 2001). Notably, dec-RVKR-cmk has been found to block SARS-CoV-2 S by inhibiting syncytium formation and virus infection with an IC50 of 5 μM (Hoffmann et al. 2020b). Bestle and co-workers evaluated the peptidomimetic furin inhibitory action of MI-1851 by western blotting and found that MI-1851 prevented SARS-CoV-2 S protein cleavage (Bestle et al. 2020).
Peptides Targeting TMPRSS2
TMPRSS2 is a serine protease that is widespread in epithelial cells of the respiratory, gastrointestinal, and urogenital tracts and is involved in the proteolytic activation of many respiratory viruses such as SARS-CoV-1 and SARS-CoV-2 (Iwata-Yoshikawa et al. 2019). Bestle and co-workers analyzed the inhibitory activity of the polypeptide aprotinin by invitro. They found it to suppress the virus production by 25 to 100-fold at a concentration of 10 μM at 16–48 h post infection (Bestle et al. 2020). They also demonstrated that the peptidomimetic inhibitor MI-432 and its analog MI-1900 are more potent than aprotinin (Meyer et al. 2013).
Peptides Targeting Cathepsin L
In the pH ranges of 3–3.5, it is observed that the SARS-CoV-1 and -2 S proteins are being activated by the cathepsin L proteases, which are widely expressed in endosomes and lysosomes. Hence the inhibitory activity could be achieved by agents that can increase the pH or decrease acidification (Simmons et al. 2005; Bosch et al. 2008; Ou et al. 2020). In this way, a peptide-P9, derived from mouse β-defensin-4, is active in inhibiting the cathepsin-L by inhibiting endosome acidification. P9 was further modified with proton accepting amino acids, resulting in peptide P9R, which is more involved than P9 against MERS-CoV, SARS-CoV-1, and SARS-CoV-2 with IC50 values in the low μg/ml range. Researchers have also found that an eight-branched derivative of P9R named 8P9R is recognized to exhibit the dual antiviral activity of inhibiting both endosome acidification (Cathepsin-L) and infection of TMPRSS2 positive Calu-3 cells by aggregating viral particles (Zhao et al. 2020, 2021). In another study by Zhang and co-workers, a glycopeptide antibiotic, teicoplanin, is also an active candidate for inhibiting cathepsin-L (Mukhopadhyay 2014).
Design of New Peptide Inhibitors
It is a proven strategy that the in silico studies for screening various peptide-based inhibitors followed by the development of potential candidates is a promising approach (Ojha et al. 2021). The entry of SARS-COV-2 in human cells occurs with the interaction of viral S protein with the ACE2 receptors of human cells (Fig. 2). The first coordinates of these complex interactions are now available in the protein data bank under the IDs: 6VW1 (Shang et al. 2020), 6M17 (Yan et al. 2020), 6lZG (Wang et al. 2020), and 6M0J (Lan et al. 2020). So, far various in silico studies have been undergoing to extract the candidates that can bind with ACE2 receptor to inhibit the interaction with the RBD domain of SARS-CoV-2 S proteins. On the other hand, various studies are also targeting inhibiting the interactions of ACE2 receptors with the HR1 domain of S protein. An essential factor is to study the structural and dynamical properties of the target and the protein interacting with it. Molecular dynamics study of the ACE2 receptor bound to the RBDs of both SARS-CoV-1 and SARS-CoV-2 revealing specific interactions stabilizing the binding mode of the viruses was reported (Ali and Vijayan 2020). Two key glycosylation sites (N165 and N234) were identified to modulate the ratio between open and closed states of S protein based on the recently full-length model of the S protein (Casalino et al. 2020).
Various necessary computational tools exploring the peptides libraries of interest are homology modeling, computational mutagenesis, docking protocols, re-scoring methods, and molecular dynamics simulations. Molecular dynamical simulations were performed with the full-atom resolution and coarse-grained models in the microseconds time scale of the SARS-CoV-2 proteome by the D.E. Shaw Research group and Sergio Pantano group Pasteur Institute of Montevideo (Pantano 2021; Shaw 2020; ). These studies infer the conformational events and interactions occurring in the SARS-CoV-2 virion structure, which helps develop novel peptide-based candidates to inhibit the entry of SARS COV.
EK1 (SLDQINVTFLDLEYEMKKLEEAIKKLEESYIDLKEL) is a pan-CoV fusion inhibitor designed by Xia and colleagues. It is an entry inhibiting peptide that is effective against SARS-CoV. EK1 acts by blocking the HR1 domain and disrupts the formation of the 6HB core, which inhibits viral fusion entry in the host cell. The peptide has shown prophylactic and therapeutic activity against various members of the coronavirus family (Mahendran et al. 2020). Conjugating cholesterol moiety with EK1 at the C-terminal extreme using glycine/serine as a linker or a polyethylene glycol spacer enhances the antiviral activity of the peptide. It was seen that the entry of SARS-CoV-2 by cell–cell fusion was severely impaired at 50% inhibitory concentration.
HD5 (ATCYCRTGRCATRESLSGVCEISGRLYRLCC) is a natural lectin-like human defensins-5 peptide; it binds to and shields ACE2 from viral recognition and binding. Firstly, based on the structure, it was reported to bind and inhibit the spike protein of SARS-CoV, but later it was said to block human ACE2 competitively (Mahendran et al. 2020). This indirectly acts as an entry inhibiting peptide by blocking the interaction of SARS-CoV spike protein with human ACE2, which is essential for viral fusion entry in the host cell (Sabbah et al. 2021; Schafer et al. 2021).
Recently, Hamid Madanchi and co-workers have evaluated the inhibitory effect of an FDA approved HIV-fusion inhibitor called Enfuviritide (Enf) (Ahmadi et al. 2021). Enf is a peptide-based inhibitor containing 36 amino acid residues and found to inhibit the fusion of HIV-1 and the host cell membrane by binding with the HR1 from the pre-hairpin fusion intermediate. Based on the resemblance of cell fusion mechanism of SARS-CoV-2 and HIV-1 cell fusion mechanism, Enf was analysed for SARS-CoV-2. In their study, interestingly they found that Enf has a very good interactions with the main amino acid residues of the HR2 domain of SARS-CoV-2 and could have a strong inhibitory action on the SARS-CoV-2.
It is noteworthy to mention the importance of antimicrobial and antiviral peptides. Even, AVPs can show good inhibitory actions like entry inhibition, inhibition of fusion to the target cell membrane. Some of the AVPs can interact with the receptor binding site of the virus and the sequence RLxRxMxxxK of these peptides is well known for their inhibitory action. Hamid Madanchi and co-workers have summarised the properties and relevance of these peptides for SARS-CoV-2 in their review (Mousavi Maleki et al. 2021). There are several peptide sequences under investigation for entry inhibition of SARS-CoV-2 virus (Table 5).
Conclusion and Future Prospects
This review outlined the progress of investigations related to peptide-based antibodies and therapeutics for the SARS-CoV-2 pandemic, summing up the latest and conceivable remedial procedures. The review discusses the primary introduction about the type of peptide-based therapeutics that can be explored for targeting SARS-CoV-2. Further, the expected mechanism of action of each type is described.
The paper is centered around the peptide-based vaccines, peptide immune suppressing peptides and immunomodulatory peptides and further describes primary viral proteins, which play a significant role in developing SARS-CoV-2 vaccinations in this concern. In the short term, repurposed medications and vaccines will be the top line of safeguard against COVID-19. But the side effects of vaccinations and drug resistance ought to permit space for the advancement of therapeutics that focus on different pathways. Peptide-based therapeutics presents several benefits over conventional small molecules, including ease of synthetic methods.
It is noteworthy to mention that synthetically modified peptides have excellent prospects because of their tunable properties for improving their stability and bioavailability. Peptides are magnificent lead compounds for their high selectivity regardless of their low plasma stability and oral bioavailability, which can be modified. Additionally, novel drug delivery systems could also be developed to improve their administration strategies.
References
Abdelmageed MI, Abdelmoneim AH, Mustafa MI, Elfadol NM, Murshed NS, Shantier SW, Makhawi AM (2020) Design of a multiepitope-based peptide vaccine against the E protein of human COVID-19: an immunoinformatics approach. Biomed Res Int 2020:e2683286. https://doi.org/10.1155/2020/2683286
Agrawal U, Raju R, Udwadia ZF (2020) Favipiravir: a new and emerging antiviral option in COVID-19. Med J Armed Forces India 76(4):370–376. https://doi.org/10.1016/j.mjafi.2020.08.004
Ahmadi K, Farasat A, Rostamian M, Johari B, Madanchi H (2021) Enfuvirtide, an HIV-1 fusion inhibitor peptide, can act as a potent SARS-CoV-2 fusion inhibitor: an in silico drug repurposing study. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2021.1871958
Ahmed A, Siman-Tov G, Hall G, Bhalla N, Narayanan A (2019) Human antimicrobial peptides as therapeutics for viral infections. Viruses 11(8):704. https://doi.org/10.3390/v11080704
Alam A, Khan A, Imam N, FaizanSiddiqui M, Waseem M, Malik Z, Ishrat R (2021) Design of an epitope-based peptide vaccine against the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): a vaccine-informatics approach. bioRxiv. https://doi.org/10.1101/2020.05.03.074930
Ali A, Vijayan R (2020) Dynamics of the ACE2–SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci Rep 10(1):1–12. https://doi.org/10.1038/s41598-020-71188-3
Sazgarnejad S, Yazdanpanah N, Rezaei N (2021) Anti-inflammatory effects of GLP-1 in patients with COVID-19. Expert Rev Anti Infec Ther. https://doi.org/10.1080/14787210.2021.1964955
Arch Biopartners Inc. (2021) Multicenter, randomized, double-blind, placebo-controlled, proof of concept study of LSALT peptide as prevention of acute respiratory distress syndrome (ARDS) and acute kidney injury in patients infected with SARS-CoV-2 (COVID-19); Clinical trial registration NCT04402957; clinicaltrials.gov.
Assistance Publique (2020) Assistance Publique—Hôpitaux de Paris. Cohort multiple randomized controlled trials open-label of immune modulatory drugs and other treatments in COVID-19 patients CORIMUNO-COAG Trial; Clinical trial registration NCT04344756; clinicaltrials.gov.
AstraZeneca (2021) A phase I double-blind, placebo-controlled study to evaluate the safety, tolerability and pharmacokinetics of AZD7442 in healthy adults; Clinical trial registration NCT04507256; clinicaltrials.gov.
Bar-Natan M (2021) A phase I open label peptide based vaccine in patients with myeloproliferative neoplasm harboring CALR mutations; Clinical trial registration NCT05025488; clinicaltrials.gov.
Becker GL, Sielaff F, Than ME, Lindberg I, Routhier S, Day R, Lu Y, Garten W, Steinmetzer T (2010) Potent inhibitors of furin and furin-like proprotein convertases containing decarboxylated P1 arginine mimetics. J Med Chem 53(3):1067–1075. https://doi.org/10.1021/jm9012455
Bergeron E, Vincent MJ, Wickham L, Hamelin J, Basak A, Nichol ST, Chrétien M, Seidah NG (2005) Implication of proprotein convertases in the processing and spread of severe acute respiratory syndrome coronavirus. Biochem Biophys Res Commun 326(3):554–563. https://doi.org/10.1016/j.bbrc.2004.11.063
Bestle D, Heindl MR, Limburg H, van Van Lam T, Pilgram O, Moulton H, Stein DA, Hardes K, Eickmann M, Dolnik O, Rohde C, Klenk HD, Garten W, Steinmetzer T, Böttcher-Friebertshäuser E (2020) TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 3(9):1–14. https://doi.org/10.26508/LSA.202000786
Bionor Immuno AS (2017) Re-boosting of subjects previously included in the CT BI-Vacc-4x 2007/1 Study. An open, multicenter, immunogenicity, follow-up re-boosting study with Vacc-4x in subjects infected with HIV-1 who have maintained an adequate response to ART; Clinical trial registration NCT01712256; clinicaltrials.gov.
Blank A, Markert C, Hohmann N, Carls A, Mikus G, Lehr T, Alexandrov A, Haag M, Schwab M, Urban S, Haefeli WE (2016) First-in-human application of the novel hepatitis B and hepatitis D virus entry inhibitor myrcludex B. J Hepatol 65(3):483–489. https://doi.org/10.1016/j.jhep.2016.04.013
Bosch BJ, Bartelink W, Rottier PJM (2008) Cathepsin L Functionally cleaves the severe acute respiratory syndrome coronavirus class I Fusion protein upstream of rather than adjacent to the fusion peptide. J Virol 82(17):8887–8890. https://doi.org/10.1128/jvi.00415-08
Bosnalijek DD (2020) An open-label, prospective, randomized, comparative clinical trial to evaluate the efficacy and safety of ENKORTEN® as an immunomodulatory therapy, within the usual therapeutically established protocol, for the treatment of patients with moderate to severe COVID-19 infection; Clinical trial registration NCT04374032; clinicaltrials.gov.
Braun E, Sauter D (2019) Furin-Mediated protein processing in infectious diseases and cancer. Clin Transl Immunol 8(8):1–19. https://doi.org/10.1002/cti2.1073
Bruno BJ, Miller GD, Lim CS (2013) Basics and recent advances in peptide and protein drug delivery. Ther Deliv 4(11):1443–1467. https://doi.org/10.4155/tde.13.104
Bruzzese V, Lazzarino AI (2020) The paradoxical effect of IL-6 and implications for the use of tocilizumab in Covid-19 patients. Med Hypotheses 144:110284. https://doi.org/10.1016/j.mehy.2020.110284
Cadila Healthcare Limited. (2021) A phase II, Multicenter, open-label, randomized, comparator controlled study to evaluate the efficacy and safety of pegylated interferon—Α2b in the treatment of adult patients diagnosed with SARS-CoV2 (COVID-19); Clinical trial registration NCT04480138; clinicaltrials.gov.
Cao L, Goreshnik I, Coventry B, Case JB, Miller L, Kozodoy L, Chen RE, Carter L, Walls AC, Park YJ, Strauch EM, Stewart L, Diamond MS, Veesler D, De BD (2020) Novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science 370(6515):426–431. https://doi.org/10.1126/science.abd9909
Casalino L, Gaieb Z, Goldsmith JA, Hjorth CK, Dommer AC, Harbison AM, Fogarty CA, Barros EP, Taylor BC, Mclellan JS, Fadda E, Amaro RE (2020) Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent Sci 6(10):1722–1734. https://doi.org/10.1021/acscentsci.0c01056
CENTOGENE GmbH Rostock (2021) Peptide-Based immunization for colon- and pancreas-carcinoma (PICOP-GLOBAL): an international, multicenter protocol; Clinical trial registration NCT03871790; clinicaltrials.gov.
Centre Hospitalier Universitaire de Besancon (2021) Anticancer therapeutic vaccination using telomerase-derives universal cancer peptides in metastatic non small cell lung cancer : a phase I/II study; Clinical trial registration NCT02818426; clinicaltrials.gov.
Channappanavar R, Zhao J, Perlman S (2014) T cell-mediated immune response to respiratory coronaviruses. Immunol Res 59(1–3):118–128. https://doi.org/10.1007/s12026-014-8534-z
Chatham WW (2021) Early treatment of cytokine storm syndrome in Covid-19; Clinical trial registration NCT04362111; clinicaltrials.gov.
Cliniques universitaires Saint-Luc- Université Catholique de Louvain (2020) Activity and safety of peptide-based immunotherapy in an umbrella window pre-operative opportunity phase II study in patients with squamous cell carcinoma of the head and neck.; Clinical trial registration NCT04445064; clinicaltrials.gov.
COL George Peoples, MD, FACS (2020). Phase I/IIa trial of folate binding protein (FBP) peptide (E39) vaccine in ovarian and endometrial cancer patients; Clinical trial registration NCT01580696; clinicaltrials.gov.
COVID-19 Vaccine https://vaxxinity.com/covid-19-vaccine/. Accessed 9 Sept 2021a
COVID-19 https://www.cytodyn.com/pipeline/covid-19. Accessed 9 Sept 2021b
Craig L, Slingluff Jr, et al. (2021). A multicenter trial to evaluate the effects of administration of cyclophosphamide and melanoma-derived helper peptides on the immunogenicity of a class I MHC-restricted peptide-based vaccine in participants with resected melanoma; Clinical trial registration NCT00118274; clinicaltrials.gov.
Dadar M, Shahali Y, Chakraborty S, Prasad M, Tahoori F, Tiwari R, Dhama K (2019) Antiinflammatory peptides: current knowledge and promising prospects. Inflamm Res 68(2):125–145. https://doi.org/10.1007/s00011-018-1208-x
Di Natale C, La Manna S, De Benedictis I, Brandi P, Marasco D (2020) Perspectives in peptide-based vaccination strategies for syndrome coronavirus 2 pandemic. Front Pharmacol. https://doi.org/10.3389/fphar.2020.578382
Dietrich P-Y (2016) Phase I/II study of intradermal IMA950 peptide-based vaccine adjuvanted with intra muscular poly-ICLC in combination with temozolomide in newly diagnosed HLA-A2 glioblastoma patients; Clinical trial registration NCT01920191; clinicaltrials.gov.
Disis M (Nora). Phase I/II study of adoptive T cell therapy following in vivo priming with a HER-2/Neu (HER2) intracellular domain (ICD) peptide-based vaccine in patients with advanced stage HER2 overexpressing breast cancer; Clinical trial registration NCT00791037; clinicaltrials.gov.
Dominguez-Horta M, Del C (2020) CIGB-258, An immunomodulatory peptide for the treatment of a COVID-19-associated hepatic encephalopathy: a case report
Du L, He Y, Zhou Y, Liu S, Zheng B-J, Jiang S (2009a) The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol 7(3):226–236. https://doi.org/10.1038/nrmicro2090
Du L, He Y, Zhou Y, Liu S, Zheng BJ, Jiang S (2009b) The spike protein of SARS-CoV—a target for vaccine and therapeutic development. Nat Rev Microbiol 7(3):226–236. https://doi.org/10.1038/nrmicro2090
Eggink D, Bontjer I, de Taeye SW, Langedijk JPM, Berkhout B, Sanders RW (2019) HIV-1 anchor inhibitors and membrane fusion inhibitors target distinct but overlapping steps in virus entry. J Biol Chem 294(15):5736–5746. https://doi.org/10.1074/jbc.RA119.007360
Federal Budgetary Research Institution State Research Center of Virology and Biotechnology “Vector” (2021) Multicenter double-blind placebo-controlled comparative randomized study of the tolerability, safety, immunogenicity and prophylactic efficacy of the EpiVacCorona peptide antigen-based vaccine for the prevention of COVID-19, With the participation of 3000 volunteers aged 18 years and above (phase III–IV); Clinical trial registration NCT04780035; clinicaltrials.gov.
FLOVID-20|Flow Pharma https://flowpharma.com/product/flovid-20/. Accessed 9 Sept 2021
Global anti-inflammatory peptides market size 2021 SWOT analysis, top manufacturers, type and application with Covid-19 impact till 2025h https://www.ktvn.com/story/43831183/global-anti-inflammatory-peptides-market-size-2021-swot-analysis-top-manufacturers-type-and-application-with-covid-19-impact-till-2025. Accessed 19 Oct 2021
Gorbalenya AE, Baker SC, Baric RS, de Groot RJ, Drosten C, Gulyaeva AA, Haagmans BL, Lauber C, Leontovich AM, Neuman BW, Penzar D, Perlman S, Poon LLM, Samborskiy DV, Sidorov IA, Sola I, Ziebuhr J (2020) The species severe acute respiratory syndrome-related coronavirus: classifying 2019-NCoV and naming It SARS-CoV-2. Nat Microbiol 5(4):536–544. https://doi.org/10.1038/s41564-020-0695-z
Gordon DE et al (2020) A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583(7816):459–468. https://doi.org/10.1038/s41586-020-2286-9
Gur M, Taka E, Yilmaz SZ, Kilinc C, Aktas U, Golcuk M (2020) Conformational transition of SARS-CoV-2 spike glycoprotein between its closed and open states. J Chem Phys. https://doi.org/10.1063/5.0011141
Hamming I, Timens W, Bulthuis MLC, Lely AT, Navis GJ, van Goor H (2004) Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A First step in understanding SARS pathogenesis. J Pathol 203(2):631–637. https://doi.org/10.1002/path.1570
Han DP, Penn-Nicholson A, Cho MW (2006) Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology 350(1):15–25. https://doi.org/10.1016/j.virol.2006.01.029
Herbert Irving Comprehensive Cancer Center (2013) A phase I study of peptide based vaccine therapy in patients with high-risk or metastatic melanoma; Clinical trial registration NCT00037037; clinicaltrials.gov.
Herst CV, Burkholz S, Sidney J, Sette A, Harris PE, Massey S, Brasel T, Cunha-Neto E, Rosa DS, Chao WCH, Carback R, Hodge T, Wang L, Ciotlos S, Lloyd P, Rubsamen R (2020) An Effective CTL peptide vaccine for Ebola Zaire based on survivors’ CD8+ targeting of a particular nucleocapsid protein epitope with potential implications for COVID-19 vaccine design. Vaccine 38(28):4464–4475. https://doi.org/10.1016/j.vaccine.2020.04.034
Ho TY, Wu SL, Chen JC, Wei YC, Cheng SE, Chang YH, Liu HJ, Hsiang CY (2006) Design and biological activities of novel inhibitory peptides for SARS-CoV Spike protein and angiotensin-converting enzyme 2 interaction. Antiviral Res 69(2):70–76. https://doi.org/10.1016/j.antiviral.2005.10.005
Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Müller MA, Drosten C, Pöhlmann S (2020a) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181(2):271-280.e8. https://doi.org/10.1016/j.cell.2020.02.052
Hoffmann M, Kleine-Weber H, Pöhlmann S (2020b) A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78(4):779-784.e5. https://doi.org/10.1016/j.molcel.2020.04.022
Hofmann H, Pyrc K, Van Der Hoek L, Geier M, Berkhout B, Pöhlmann S (2005) Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA 102(22):7988–7993. https://doi.org/10.1073/pnas.0409465102
Human Gamma Globulin (2021) Human gamma globulin: view uses, side effects and medicines https://www.1mg.com/generics/human-gamma-globulin-209993. Accessed 19 Oct 2021
Immatics Biotechnologies GmbH (2014) A phase 1 trial of peptide-based glioma vaccine IMA950 in patients with glioblastoma (GBM); Clinical trial registration NCT01403285; clinicaltrials.gov.
Immune modulators for treating COVID-19 (2021) ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04593940. Accessed 21 Oct 2021
Interferon Beta 1a in hospitalized COVID-19 patients (2021) ClinicalTrials.gov https://clinicaltrials.gov/ct2/show/NCT04350671. Accessed 21 Oct 2021
Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N (2019) TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. https://doi.org/10.1128/jvi.01815-18
Jr C. L. S. (2016a) Evaluation of CD8+ T cell activation and infiltration into primary breast tumors following administration of a peptide vaccine; Clinical trial registration NCT00892567; clinicaltrials.gov.
Jr C. L. S. (2016b) Evaluation of the safety and immunogenicity of intratumoral injection of interferon gamma during vaccination in patients with subcutaneous or cutaneous metastases of melanoma; Clinical trial registration NCT00977145; clinicaltrials.gov.
Jr C. L. S. (2016c) Combination therapy of topical imiquimod plus multipeptide vaccination for cutaneous metastases of melanoma; Clinical trial registration NCT01264731; clinicaltrials.gov.
Jr C. L. S. (2020) Evaluation of different adjuvants for the transdermal administration of a peptide-based vaccine in participants with high-risk melanoma; Clinical trial registration NCT00118313; clinicaltrials.gov.
Kalita P, Padhi AK, Zhang KYJ, Tripathi T (2020) Design of a peptide-based subunit vaccine against novel coronavirus SARS-CoV-2. Microb Pathog 145:104236. https://doi.org/10.1016/j.micpath.2020.104236
Katsiki N, Ferrannini E (2020) Anti-inflammatory properties of antidiabetic drugs: a “promised land” in the COVID-19 era? J Diabetes Complicat 34(12):107723. https://doi.org/10.1016/j.jdiacomp.2020.107723
Kurpe SR, Grishin SYu, Surin AK, Panfilov AV, Slizen MV, Chowdhury SD, Galzitskaya OV (2020) Antimicrobial and amyloidogenic activity of peptides. Can antimicrobial peptides be used against SARS-CoV-2? Int J Mol Sci 21(24):9552. https://doi.org/10.3390/ijms21249552
La Manna S, Di Natale C, Florio D, Marasco D (2018) Peptides as therapeutic agents for inflammatory-related diseases. Int J Mol Sci 19(9):2714. https://doi.org/10.3390/ijms19092714
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807):215–220. https://doi.org/10.1038/s41586-020-2180-5
Lee N, Heo YJ, Choi S-E, Jeon JY, Han SJ, Kim DJ, Kang Y, Lee KW, Kim HJ (2021) Anti-inflammatory effects of empagliflozin and gemigliptin on LPS-stimulated macrophage via the IKK/NF-κB, MKK7/JNK, and JAK2/STAT1 signalling pathways. J Immunol Res 2021:9944880. https://doi.org/10.1155/2021/9944880
Li Q et al (2020) Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med 382(13):1199–1207. https://doi.org/10.1056/NEJMoa2001316
Liu Y, Pan Y, Hu Z, Wu M, Wang C, Feng Z, Mao C, Tan Y, Liu Y, Chen L, Li M, Wang G, Yuan Z, Diao B, Wu Y, Chen Y (2020) Thymosin alpha 1 reduces the mortality of severe coronavirus disease 2019 by restoration of lymphocytopenia and reversion of exhausted T cells. Clin Infect Dis 71(16):2150–2157. https://doi.org/10.1093/cid/ciaa630
Mahendran ASK, Lim YS, Fang C-M, Loh H-S, Le CF (2020) The potential of antiviral peptides as COVID-19 therapeutics. Front Pharmacol 11:1475. https://doi.org/10.3389/fphar.2020.575444
Mahmudpour M, Roozbeh J, Keshavarz M, Farrokhi S, Nabipour I (2020) COVID-19 cytokine storm: the anger of inflammation. Cytokine 133:155151. https://doi.org/10.1016/j.cyto.2020.155151
Mayo Clinic (2018) MUC1/HER-2/Neu peptide based immunotherapeutic vaccines for breast adenocarcinomas; Clinical trial registration NCT00640861; clinicaltrials.gov.
Meyer D, Sielaff F, Hammami M, Böttcher-Friebertshäuser E, Garten W, Steinmetzer T (2013) Identification of the first synthetic inhibitors of the type II transmembrane serine protease TMPRSS2 suitable for inhibition of influenza virus activation. Biochem J 452(2):331–343. https://doi.org/10.1042/BJ20130101
Mohamed AMS (2020) Randomized prospective clinical study for efficacy of anti-corona VS2 immunoglobulins prepared from COVID19 convalescent plasma in prevention of infection in high risk groups as well as treatment of early cases of COVID19 patients; Clinical trial registration NCT04383548; clinicaltrials.gov.
Mousavi Maleki MS, Rostamian M, Madanchi H (2021) Antimicrobial peptides and other peptide-like therapeutics as promising candidates to combat SARS-CoV-2. Expert Rev Anti Infect Ther 19(10):1205–1217. https://doi.org/10.1080/14787210.2021.1912593
Mukhopadhyay J (2014) Teicoplanin for treating MRSA pneumonia. BMJ (online) 348:2317. https://doi.org/10.1136/bmj.g2317
Muttenthaler M, King GF, Adams DJ, Alewood PF (2021) Trends in peptide drug discovery. Nat Rev Drug Discov 20(4):309–325. https://doi.org/10.1038/s41573-020-00135-8
Nevagi RJ, Toth I, Skwarczynski M (2018) Peptide-based vaccines. In: Koutsopoulos S (ed) Peptide applications in biomedicine, biotechnology and bioengineering. Woodhead Publishing, Sawston
Novartis Pharmaceuticals (2021) Novartis pharmaceuticals phase 3 multicenter, randomized, double-blind, placebo-controlled study to assess the efficacy and safety of canakinumab on cytokine release syndrome in patients with COVID-19-induced pneumonia (CAN-COVID); Clinical trial registration NCT04362813; clinicaltrials.gov.
Nyanguile O (2019) Peptide antiviral strategies as an alternative to treat lower respiratory viral infections. Front Immunol 10:1366. https://doi.org/10.3389/fimmu.2019.01366
Octapharma (2021) Multinational, observational, retrospective study to evaluate coagulation changes and the influence of antithrombin III treatment in patients with severe COVID-19 infection; Clinical trial registration NCT04651400; clinicaltrials.gov.
Ojha PK, Kar S, Krishna JG, Roy K, Leszczynski J (2021) Therapeutics for COVID-19: from computation to practices—where we are, where we are heading to, vol 25. Springer, Cham
OSE Immunotherapeutics (2021) A randomized, open label, phase 1 study to evaluate the safety, reactogenicity and immunogenicity of OSE-13E, a multiepitope-based vaccine candidate against COVID-19, in healthy adults (COVEPIT-3); Clinical trial registration NCT04885361; clinicaltrials.gov.
Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z (2020) Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. https://doi.org/10.1038/s41467-020-15562-9
Peptide drug discovery could lead to a powerful anti-inflammatory https://www.genengnews.com/news/peptide-drug-discovery-could-lead-to-a-powerful-anti-inflammatory/. Accessed 19 Oct 2021
Poh CM et al (2020) Two linear epitopes on the SARS-CoV-2 Spike protein that elicit neutralising antibodies in COVID-19 patients. Nat Commun 11(1):2806. https://doi.org/10.1038/s41467-020-16638-2
Poor H (2021) Tenecteplase with concomitant anticoagulation for severe acute respiratory failure in patients with COVID-19; Clinical trial registration NCT04505592; clinicaltrials.gov.
Pouwels KB, Pritchard E, Matthews PC et al (2021) Effect of Delta variant on viral burden and vaccine effectiveness against new SARS-CoV-2 infections in the UK. Nat Med 27:2127–2135. https://doi.org/10.1038/s41591-021-01548-7
Prachar M, Justesen S, Steen-Jensen DB, Thorgrimsen S, Jurgons E, Winther O, Bagger FO (2020) COVID-19 vaccine candidates prediction and validation of 174 SARS-CoV-2 epitopes. bioRxiv. https://doi.org/10.1101/2020.03.20.000794
Purcell AW, McCluskey J, Rossjohn J (2007) More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov 6(5):404–414. https://doi.org/10.1038/nrd2224
Raby B (2021) Pulmozyme to improve COVID-19 ARDS outcomes; Clinical trial registration NCT04402944; clinicaltrials.gov.
Revimmune (2021) Recombinant human interleukin-7 (CYT107) to improve clinical outcomes in lymphopenic patients with COVID-19 infection—“ILIAD 7 Trial” Brazil Cohort; Clinical trial registration NCT04927169; clinicaltrials.gov.
Rhode Island Hospital (2021) A pilot trial of thymalfasin (Ta1) to treat COVID-19 infection in patients with lymphocytopenia; Clinical trial registration NCT04487444; clinicaltrials.gov.
Robbiani DF et al (2020) Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584(7821):437–442. https://doi.org/10.1038/s41586-020-2456-9
Roshanravan N, Seif F, Ostadrahimi A, Pouraghaei M, Ghaffari S (2020) Targeting cytokine storm to manage patients with COVID-19: a mini-review. Arch Med Res 51(7):608–612. https://doi.org/10.1016/j.arcmed.2020.06.012
Ruscitti P, Berardicurti O, Iagnocco A, Giacomelli R (2020) Cytokine storm syndrome in severe COVID-19. Autoimmun Rev 19(7):102562. https://doi.org/10.1016/j.autrev.2020.102562
Sabbah DA, Hajjo R, Bardaweel SK, Zhong HA (2021) An updated review on betacoronavirus viral entry inhibitors: learning from past discoveries to advance COVID-19 drug discovery. Curr Top Med Chem 21(7):571–596. https://doi.org/10.2174/1568026621666210119111409
Sala A, Ardizzoni A, Ciociola T, Magliani W, Conti S, Blasi E, Cermelli C (2018) Antiviral activity of synthetic peptides derived from physiological proteins. Intervirology 61(4):166–173
Schafer A, Xiong R, Cooper L, Nowar R, Lee H, Li Y, Ramirez BE, Peet NP, Caffrey M, Thatcher GR, Saphire EO (2021) Evidence for distinct mechanisms of small molecule inhibitors of filovirus entry. PLoS Pathog 17(2):e1009312
Sergio Pantano (2021) Biochemical and Biophysical Research Communications. Elsevier, Hoboken. https://www.journals.elsevier.com/biochemical-and-biophysical-research-communications/editorial-board/sergio-pantano. Accessed 19 Oct 2021
Shahid Beheshti University of Medical Sciences (2020) The clinical trial of application of ezrin peptide (HEP-1) for treatment of coronavirus disease (COVID-19) infection; Clinical trial registration NCT04627233; clinicaltrials.gov.
Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F (2020) Structural basis of receptor recognition by SARS-CoV-2. Nature 581(7807):221–224. https://doi.org/10.1038/s41586-020-2179-y
Shaw DE (2020) Molecular dynamics simulations related to Sars-Cov-2. DE Shaw Research Technical Data
Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P (2005) Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA 102(33):11876–11881. https://doi.org/10.1073/pnas.0505577102
Southwest Oncology Group (2015) An early phase study of an EGFRvIII peptide based vaccine in patients with EGFRvIII expressing cancers; Clinical trial registration NCT00023634; clinicaltrials.gov.
Soy M, Keser G, Atagündüz P, Tabak F, Atagündüz I, Kayhan S (2020) Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin Rheumatol 39(7):2085–2094. https://doi.org/10.1007/s10067-020-05190-5
Sugrue RJ, Brown C, Brown G, Aitken J, McL. Rixon HW (2001) Furin cleavage of the respiratory syncytial virus fusion protein is not a requirement for its transport to the surface of virus-infected cells. J Gen Virol 82(6):1375–1386. https://doi.org/10.1099/0022-1317-82-6-1375
Taylor PC, Adams AC, Hufford MM, de la Torre I, Winthrop K, Gottlieb RL (2021) Neutralizing monoclonal antibodies for treatment of COVID-19. Nat Rev Immunol 21(6):382–393. https://doi.org/10.1038/s41577-021-00542-x
Testing for COVID-19: a way to lift confinement restrictions https://www.oecd.org/coronavirus/policy-responses/testing-for-covid-19-a-way-to-lift-confinement-restrictions-89756248/. Accessed 6 Jan 2022
Tizzano B, Palladino P, De Capua A, Marasco D, Rossi F, Benedetti E, Pedone C, Ragone R, Ruvo M (2005) The human prion protein Α2 helix: a thermodynamic study of its conformational preferences. Proteins 59(1):72–79. https://doi.org/10.1002/prot.20395
Together against Alzheimer’s disease since 1999 http://www.axon-neuroscience.eu/. Accessed 9 Sept 2021
United Biomedical Inc., Asia. (2021) A phase I, open-label study to evaluate the safety, tolerability, and immunogenicity of UB-612 vaccine in healthy adult volunteers; Clinical trial registration NCT04545749; clinicaltrials.gov.
United Neuroscience Ltd. (2020) A phase 1 study to evaluate the safety, tolerability, and immunogenicity of UBITh® PD immunotherapeutic vaccine (UB-312) in healthy participants and participants with Parkinson’s disease; Clinical trial registration NCT04075318; clinicaltrials.gov.
University Hospital Inselspital, Berne. (2020) CORON-ACT—a multicenter, double-blind, randomized controlled phase II Trial on the efficacy and safety of tocilizumab in the treatment of Coronavirus Induced Disease (COVID-19); Clinical trial registration NCT04335071; clinicaltrials.gov.
University Hospital Tuebingen (2021a) B-PVAC-SARS-CoV-2: phase I/II multicenter safety and immunogenicity trial of multi-peptide vaccination to prevent COVID-19 infection in adults with bcell/antibody deficiency; Clinical trial registration NCT04954469; clinicaltrials.gov.
University Hospital Tuebingen (2021b) P-PVAC-SARS-CoV-2: phase I single-center safety and immunogenicity trial of multi-peptide vaccination to prevent COVID-19 infection in adults; Clinical trial registration NCT04546841; clinicaltrials.gov.
University of Southern California (2014) An open label study of MDX-CTLA4 in combination with tyrosinase/Gp100/MART-1 peptides emulsified with montanide ISA 51 in the treatment of patients with resected stage III or IV Melanoma; Clinical trial registration NCT00028431; clinicaltrials.gov.
University of Washington (2017) Phase I/II randomized study of combination immunotherapy with or without polysaccharide krestin (PSK®) concurrently with a HER2 ICD peptide-based vaccine in patients with stage IV breast cancer receiving HER2-targeted monoclonal antibody therapy; Clinical trial registration NCT01922921; clinicaltrials.gov.
University of Washington (2019) A phase I study of a HER-2/Neu peptide based vaccine with GM-CSF as an adjuvant in patients with advanced stage HER-2/Neu expressing cancers; Clinical trial registration NCT00003002; clinicaltrials.gov.
University of Washington (2020) phase II study of a HER-2/Neu (HER2) intracellular domain (ICD) peptide-based vaccine administered to patients with locally advanced or stage IV HER2 positive breast cancer; Clinical trial registration NCT00343109; clinicaltrials.gov.
Vagner J, Qu H, Hruby VJ (2008) Peptidomimetics, a synthetic tool of drug discovery. Curr Opin Chem Biol 12(3):292–296. https://doi.org/10.1016/j.cbpa.2008.03.009
Vargas MH (2020) Controlled and randomized clinical trial for evaluating the effect of a supplement of glycine as adjuvant in the treatment of COVID-19 pneumonia in patients initiating mechanical ventilation; Clinical trial registration NCT04443673; clinicaltrials.gov.
Verdecchia P, Cavallini C, Spanevello A, Angeli F (2020) The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med 76:14–20. https://doi.org/10.1016/j.ejim.2020.04.037
Vir Biotechnology|A world without infectious disease https://www.vir.bio/. Accessed 9 Sept 2021
Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, Wang Q, Zhou H, Yan J, Qi J (2020) Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181(4):894-904.e9. https://doi.org/10.1016/j.cell.2020.03.045
Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367(6485):1444–1448. https://doi.org/10.1126/science.abb2762
Yi Y, Lagniton PNP, Ye S, Li E, Xu R-H (2020) COVID-19: what has been learned and to be learned about the novel coronavirus disease. Int J Biol Sci 16(10):1753–1766. https://doi.org/10.7150/ijbs.45134
Zaldívar HMR (2021) A single-arm, open-label, phase II Clinical study to evaluate the safety and efficacy of thymic peptides in the treatment of hospitalized COVID-19 patients in honduras; Clinical trial registration NCT04771013; clinicaltrials.gov.
Zhang Y, Chen Y, Meng Z (2020) Immunomodulation for severe COVID-19 pneumonia: the state of the art. Front Immunol 11:2782. https://doi.org/10.3389/fimmu.2020.577442
Zhao H, To KKW, Sze KH, Yung TTM, Bian M, Lam H, Yeung ML, Li C, Chu H, Yuen KY (2020) A broad-spectrum virus- and host-targeting peptide against respiratory viruses including influenza virus and SARS-CoV-2. Nat Commun. https://doi.org/10.1038/s41467-020-17986-9
Zhao H, To KKW, Lam H, Zhou X, Chan JFW, Peng Z, Lee ACY, Cai J, Chan WM, Ip JD, Chan CCS, Yeung ML, Zhang AJ, Chu AWH, Jiang S, Yuen KY (2021) Cross-Linking peptide and repurposed drugs inhibit both entry pathways of SARS-CoV-2. Nat Commun. https://doi.org/10.1038/s41467-021-21825-w
Zhou Y, Fu B, Zheng X, Wang D, Zhao C, Qi Y, Sun R, Tian Z, Xu X, Wei H (2020) Pathogenic T cells and inflammatory monocytes incite inflammatory storm in severe COVID-19 patients. Natl Sci Rev. https://doi.org/10.1093/nsr/nwaa041
Author information
Authors and Affiliations
Contributions
All authors have contributed equally.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bagwe, P.V., Bagwe, P.V., Ponugoti, S.S. et al. Peptide-Based Vaccines and Therapeutics for COVID-19. Int J Pept Res Ther 28, 94 (2022). https://doi.org/10.1007/s10989-022-10397-y
Accepted:
Published:
DOI: https://doi.org/10.1007/s10989-022-10397-y