Abstract
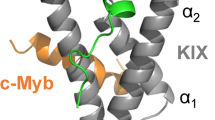
The transcriptional enhanced associate domain (TEAD) transcription factor is a core regulator of Hippo signaling pathway and has been recognized as a promising therapeutic target against gastrointestinal tumors. The intermolecular interaction of TEAD with its transcription coactivator vestigial-like protein (VGLL) is involved in the tumorigenesis of esophageal cancer, which possesses an additional double-stranded β-sheet (DSβS) motif in its TEAD-recognition domain as compared to the classic coactivators YAP and TAZ. Here, the DSβS motif is examined systematically based on the complex crystal structure of TEAD with VGL. It is found that the DSβS is one of the most important recognition sites at TEAD–VGLL complex interface, which can confer stability and specificity to the complex recognition and elicit the transcription activation event. Protein segments with different lengths are derived from the DSβS motif of VGLL to define a number of linear peptides; their binding modes and affinities to TEAD are computationally analyzed to reveal that these linear peptides cannot maintain in native double-strand conformation in free solution state, thus unfavorable for their rebinding to TEAD–VGLL interface. A head-to-tail cyclization strategy is described to constrain the conformation of linear peptides, which can considerably reduce the peptide flexibility and help the peptide folding to an approximately native conformation in solution. We also demonstrate that the binding affinity and biological activity of cyclic peptides are improved effectively relative to their linear counterparts at molecular and cellular levels.




Similar content being viewed by others
References
Boopathy GTK, Hong W (2019) Role of hippo pathway-YAP/TAZ signaling in angiogenesis. Front Cell Dev Biol 7:49
Choi W, Kim J, Park J, Lee DH, Hwang D, Kim JH, Ashktorab H, Smoot D, Kim SY, Choi C, Koh GY, Lim DS (2018) YAP/TAZ initiates gastric tumorigenesis via upregulation of MYC. Cancer Res 78:3306–3320
Furet P, Salem B, Mesrouze T, Schmelzle T, Lewis I, Kallen J, Chène P (2019) Structure-based design of potent linear peptide inhibitors of the YAP–TEAD protein–protein interaction derived from the YAP Ω-loop sequence. Bioorg Med Chem Lett 29(16):2316–2319
Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, Onufriev A (2005) H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res 33:W368–W371
Han M, Sun D (2019) Rational creation and systematic analysis of cervical cancer kinase–inhibitor binding profile. J Comput Aided Mol Des 33:689–698
Hekkelman ML, Te Beek TAH, Pettifer SR, Thorne D, Attwood TK, Vriend G (2010) WIWS: a protein structure bioinformatics Web service collection. Nucleic Acids Res 38:W719–W723
Holden JK, Cunningham CM (2018) Targeting the Hippo pathway and cancer through the TEAD family of transcription factors. Cancers 10:81
Jiang W, Yao F, He J, Lv B, Fang W, Zhu W, He G, Chen J, He J (2015) Downregulation of VGLL4 in the progression of esophageal squamous cell carcinoma. Tumour Biol 36:1289–1297
Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, Wang W, Wang X, Guo T, Li P, Zhao Y, Ji H, Zhang L, Zhou Z (2014) A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25:166–180
Kang W, Cheng ASL, Yu J, To KF (2016) Emerging role of Hippo pathway in gastric and other gastrointestinal cancers. World J Gastroenterol 22:1279–1288
Li Y, Roy A, Zhang Y (2009) HAAD: A quick algorithm for accurate prediction of hydrogen atoms in protein structures. PLoS ONE 4:e6701
López-Blanco JR, Canosa-Valls AJ, Li Y, Chacón P (2016) RCD+: fast loop modeling server. Nucleic Acids Res 44:W395–W400
Maupetit J, Derreumaux P, Tuffery P (2009) PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res 37:W498–W503
Mesrouze Y, Bokhovchuk F, Izaac A, Meyerhofer M, Zimmermann C, Fontana P, Schmelzle T, Erdmann D, Furet P, Kallen J, Chène P (2018) Adaptation of the bound intrinsically disordered protein YAP to mutations at the YAP:TEAD interface. Protein Sci 27:1810–1820
Michod D, Widmann C (2007) TAT-RasGAP317-326 requires p53 and PUMA to sensitize tumor cells to genotoxins. Mol Cancer Res 5:497–507
Pobbati AV, Hong W (2013) Emerging roles of TEAD transcription factors and its coactivators in cancers. Cancer Biol Ther 14:390–398
Pobbati AV, Chan SW, Lee I, Song H, Hong W (2012) Structural and functional similarity between the Vgll1–TEAD and the YAP–TEAD complexes. Structure 20:1135–1140
Raveh B, London N, Schueler-Furman O (2010) Sub-angstrom modeling of complexes between flexible peptides and globular proteins. Proteins 78:2029–2040
Spiliotopoulos D, Kastritis PL, Melquiond AS, Bonvin AM, Musco G, Rocchia W, Spitaleri A (2016) dMM-PBSA: a new HADDOCK scoring function for protein–peptide docking. Front Mol Biosci 3:46
Weng G, Gao J, Wang Z, Wang E, Hu X, Yao X, Cao D, Hou T (2020) Comprehensive evaluation of fourteen docking programs on protein–peptide complexes. J Chem Theory Comput 16:3959–3969
Wu D, Luo L, Yang Z, Chen Y, Quan Y, Min Z (2020) Targeting human Hippo TEAD binding interface with YAP/TAZ-derived, flexibility-reduced peptides in gastric cancer. Int J Pept Res Ther. https://doi.org/10.1007/s10989-020-10069-9
Yu H, Zhou P, Deng M, Shang Z (2014) Indirect readout in protein–peptide recognition: a different story from classical biomolecular recognition. J Chem Inf Model 54:2022–2032
Zhao B, Tumaneng K, Guan KL (2011) The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol 13:877–883
Zhao L, Huang Q, Tian S, Ge J, Zhu H, Dong Q (2019) Integrative identification of unexpected kinase-inhibitor interactions in the MAPK-mediated proliferation and differentiation of Mc3T3-E1 osteoblasts. Gen Physiol Biophys 38:1–13
Zhou P, Tian F, Shang Z (2009) 2D depiction of nonbonding interactions for protein complexes. J Comput Chem 30:940–951
Zhou P, Jin B, Li H, Huang SY (2018) HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res 46:W443–W450
Zhou P, Miao Q, Yan F, Li Z, Jiang Q, Wen L, Meng Y (2019) Is protein context responsible for peptide-mediated interactions? Mol Omics 15:280–295
Acknowledgements
This work was supported by the Xiamen Science and Technology Project (No. 3502Z20184050).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zheng, W., Lan, J., Feng, L. et al. Structure-Based Optimization of Conformationally Constrained Peptides to Target Esophageal Cancer TEAD Transcription Factor. Int J Pept Res Ther 27, 923–930 (2021). https://doi.org/10.1007/s10989-020-10138-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-020-10138-z