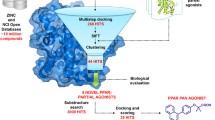
Abstract
The small-molecule modulators (agonists and antagonists) of peroxisome proliferator-activated receptor-γ (PPARγ) have been widely used to treat metabolic, cardiovascular and inflammatory diseases such as Type-2 diabetes and heart failure. Existing PPARγ modulators are all developed to target the ligand-binding site (LBS) in PPARγ ligand-binding domain (LBD) interior, where is spatially separated from the coactivator-interacting site (CIS) on PPARγ LBD surface and can thus only indirectly mediate the binding of NCOA1 coactivator peptide to CIS site. Here, we propose a new therapeutic strategy termed as Type-III PPARγ antagonists, which competitively disrupt the NCOA1 peptide binding by directly targeting PPARγ CIS site. Such antagonists are also known as inverse agonists that can partially or fully repress the basal transcriptional activity of PPARγ. A structure-based stepwise screening is performed against a purchasable biogenic compound library to identify CIS-specific binding competitors. The screening integrates empirical manual exclusion, druglikeness/pharmacokinetic evaluation, chemical similarity analysis and high-throughput molecular docking to rank the relative binding capability for compound candidate lists. The competitive potency of eight promising hits against a truncated version (core binding sequence) of full-length NCOA1 peptide (685ERHKILHRLLQEGSPS700) is tested using fluorescence competition assays. Consequently, two out of the eight tested compounds are determined as potential competitors with NCOA1 peptide for PPARγ CIS site, with CC50 values of 13 ± 1.8 and 54 ± 6.7 μM, respectively. Structural analysis reveals that the two competitor ligands are anchored at the central hole of CIS site through their substituted phenyl moieties, while forming a variety of hydrogen bonds, cation-π stacking and hydrophobic contacts with the regions surrounding the hole, conferring both stability and specificity to the CIS–competitor recognition and interaction.





Similar content being viewed by others
References
Bai Z, Hou S, Zhang S, Li Z, Zhou P (2017) Targeting self-binding peptides as a novel strategy to regulate protein activity and function: a case study on the proto-oncogene tyrosine protein kinase c-Src. J Chem Inf Model 57:835–845
Berger J, Moller DE (2002) The mechanisms of action of PPARs. Annu Rev Med 53:409–435
Butina D (1999) Unsupervised data base clustering based on daylight’s fingerprint and Tanimoto similarity: a fast and automated way to cluster small and large data sets. J Chem Inf Comput Sci 39:747–750
Chen R, Wan J, Song J, Qian Y, Liu Y, Gu S (2017) Rational screening of peroxisome proliferator-activated receptor-γ agonists from natural products: potential therapeutics for heart failure. Pharm Biol 55:503–509
Choi SS, Park J, Choi JH (2014) Revisiting PPARγ as a target for the treatment of metabolic disorders. BMB Rep 47:599–608
Choi SS, Kim ES, Jung JE, Marciano DP, Jo A, Koo JY, Choi SY, Yang YR, Jang HJ, Kim EK, Park J, Kwon HM, Lee IH, Park SB, Myung KJ, Suh PG, Griffin PR, Choi JH (2016) PPARγ antagonist Gleevec improves insulin sensitivity and promotes the browning of white adipose tissue. Diabetes 65:829–839
Doshi LS, Brahma MK, Bahirat UA, Dixit AV, Nemmani KV (2010) Discovery and development of selective PPARγ modulators as safe and effective antidiabetic agents. Expert Opin Investig Drugs 19:489–512
Guan Y (2004) Peroxisome proliferator-activated receptor family and its relationship to renal complications of the metabolic syndrome. J Am Soc Nephrol 15:2801–2815
Guasch L, Sala E, Castell-Auví A, Cedó L, Liedl KR, Wolber G, Muehlbacher M, Mulero M, Pinent M, Ardévol A, Valls C, Pujadas G, Garcia-Vallvé S (2012) Identification of PPARγ partial agonists of natural origin (I): development of a virtual screening procedure and in vitro validation. PLoS ONE 7:e50816
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45:177–182
Irwin JJ, Shoichet BK, Mysinger MM, Huang N, Colizzi F, Wassam P, Cao Y (2009) Automated docking screens: a feasibility study. J Med Chem 52:5712–5720
Jain S, Pulikuri S, Zhu Y, Qi C, Kanwar YS, Yeldandi AV, Rao MS, Reddy JK (1998) Differential expression of the peroxisome proliferator-activated receptor gamma (PPARγ) and its coactivators steroid receptor coactivator-1 and PPAR-binding protein PBP in the brown fat, urinary bladder, colon, and breast of the mouse. Am J Pathol 153:349–354
Jang JY, Koh M, Bae H, An DR, Im HN, Kim HS, Yoon JY, Yoon HJ, Han BW, Park SB, Suh SW (2017) Structural basis for differential activities of enantiomeric PPARγ agonists: binding of S35 to the alternate site. Biochim Biophys Acta 1865:674–681
Kawai M, Rosen CJ (2010) PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nat Rev Endocrinol 6:629–636
Lee SK, Chang GS, Lee IH (2004) The PreADME: PC-based program for batch predication of ADME properties. EuroQSAR 9:5–10
Lin J, Sahakian DC, de Morais SM (2003) The role of absorption, distribution, metabolism, excretion and toxicity in drug discovery. Curr Top Med Chem 3:1125–1154
Lipinski CA, Lombardo F, Dominy BW (2007) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46:3–26
Luo H, Du T, Zhou P, Yang L, Mei H, Ng H, Zhang W, Shu M, Tong W, Shi L, Mendrick DL, Hong H (2015) Molecular docking to identify associations between drugs and class I human leukocyte antigens for predicting idiosyncratic drug reactions. Comb Chem High Throughput Screen 18:296–304
Martin YC, Kofron JL, Traphagen LM (2002) Do structurally similar molecules have similar biological activity? J Med Chem 45:4350–4358
Mysinger MM, Shoichet BK (2010) Rapid context-dependent ligand desolvation in molecular docking. J Chem Inf Model 50:1561–1573
Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature 395:137–143
Ohashi M, Gamo K, Tanaka Y, Waki M, Beniyama Y, Matsuno K, Wada J, Tenta M, Eguchi J, Makishima M, Matsuura N, Oyama T, Miyachi H (2015) Structural design and synthesis of arylalkynyl amide-type peroxisome proliferator-activated receptor γ (PPARγ)-selective antagonists based on the helix12-folding inhibition hypothesis. Eur J Med Chem 90:53–67
Rieusset J, Touri F, Michalik L, Escher P, Desvergne B, Niesor E, Wahli W (2002) A new selective peroxisome proliferator-activated receptor γ antagonist with antiobesity and antidiabetic activity. Mol Endocrinol 16:2628–2644
Roehrl MH, Wang JY, Wagner G (2004) Discovery of small-molecule inhibitors of the NFAT—calcineurin interaction by competitive high-throughput fluorescence polarization screening. Biochemistry 43:16067–16075
Sood V, Colleran K, Burge MR (2000) Thiazolidinediones: a comparative review of approved uses. Diab Technol Ther 2:429–440
Tian F, Lv Y, Zhou P, Yang L (2011) Characterization of PDZ domain-peptide interactions using an integrated protocol of QM/MM, PB/SA, and CFEA analyses. J Comput Aided Mol Des 25:947–958
Tian F, Tan R, Guo T, Zhou P, Yang L (2013) Fast and reliable prediction of domain-peptide binding affinity using coarse-grained structure models. Biosystems 113:40–49
Tian F, Yang C, Wang C, Guo T, Zhou P (2014) Mutatomics analysis of the systematic thermostability profile of Bacillus subtilis lipase A. J Mol Model 20:2257
Tyagi S, Gupta P, Saini AS, Kaushal C, Sharma S (2011) The peroxisome proliferator-activated receptor: a family of nuclear receptors role in various diseases. J Adv Pharm Technol Res 2:236–240
Wang L, Waltenberger B, Pferschy-Wenzig EM, Blunder M, Liu X, Malainer C, Blazevic T, Schwaiger S, Rollinger JM, Heiss EH, Schuster D, Kopp B, Bauer R, Stuppner H, Dirsch VM, Atanasov AG (2014) Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARγ): a review. Biochem Pharmacol 92:73–89
Willson TM, Lambert MH, Kliewer SA (2001) Peroxisome proliferator-activated receptor γ and metabolic disease. Annu Rev Biochem 70:341–367
Yang C, Wang C, Zhang S, Huang J, Zhou P (2015a) Structural and energetic insights into the intermolecular interaction among human leukocyte antigens, clinical hypersensitive drugs and antigenic peptides. Mol Simul 41:741–751
Yang C, Zhang S, He P, Wang C, Huang J, Zhou P (2015b) Self-binding peptides: folding or binding. J Chem Inf Model 55:329–342
Yang C, Zhang S, Bai Z, Hou S, Wu D, Huang J, Zhou P (2016) A two-step binding mechanism for the self-binding peptide recognition of target domains. Mol Biosyst 12:1201–1213
Yu H, Zhou P, Deng M, Shang Z (2014) Indirect readout in protein–peptide recognition: a different story from classical biomolecular recognition. J Chem Inf Model 54:2022–2032
Zhang X, Pickin KA, Bose R, Jura N, Cole PA, Kuriyan J (2007) Inhibition of the EGF receptor by binding of MIG6 to an activating kinase domain interface. Nature 450:741–744
Zhang Y, Gu M, Cai W, Yu L, Feng L, Zhang L, Zang Q, Wang Y, Wang D, Chen H, Tong Q, Ji G, Huang C (2016) Dietary component isorhamnetin is a PPARγ antagonist and ameliorates metabolic disorders induced by diet or leptin deficiency. Sci Rep 6:19288
Zhang GL, Liu FY, Zhang J, Wang LP. Jia EX, Lv SM (2018) Integrated in silico-in vitro screening of ovarian cancer peroxisome proliferator-activated receptor-γ agonists against a biogenic compound library. Med Chem Res 27:341–349
Zheng W, Qiu L, Wang R, Feng X, Han Y, Zhu Y, Chen D, Liu Y, Jin L, Li Y (2015) Selective targeting of PPARγ by the natural product chelerythrine with a unique binding mode and improved antidiabetic potency. Sci Rep 5:12222
Zhou P, Yang C, Ren Y, Wang C, Tian F (2013a) What are the ideal properties for functional food peptides with antihypertensive effect? A computational peptidology approach. Food Chem 141:2967–2973
Zhou P, Wang C, Ren Y, Yang C, Tian F (2013b) Computational peptidology: a new and promising approach to therapeutic peptide design. Curr Med Chem 20:1985–1996
Zhou P, Wang C, Tian F, Ren Y, Yang C, Huang J (2013c) Biomacromolecular quantitative structure-activity relationship (BioQSAR): a proof-of-concept study on the modeling, prediction and interpretation of protein-protein binding affinity. J Comput Aided Mol Des 27:67–78
Zhou P, Zhang S, Wang Y, Yang C, Huang J (2016) Structural modeling of HLA-B1502 peptide carbamazepine T-cell receptor complex architecture: implication for the molecular mechanism of carbamazepine-induced Stevens-Johnson syndrome toxic epidermal necrolysis. J Biomol Struct Dyn 34:1806–1817
Zhou P, Hou S, Bai Z, Li Z, Wang H, Chen Z, Meng Y (2018) Disrupting the intramolecular interaction between proto-oncogene c-Src SH3 domain and its self-binding peptide PPII with rationally designed peptide ligands. Artif Cells Nanomed Biotechnol 46:1122–1131
Zieleniak A, Wójcik M, Woźniak LA (2008) Structure and physiological functions of the human peroxisome proliferator-activated receptor γ. Arch Immunol Ther Exp 56:331–345
Funding
This work was supported by the Medicine and Health Project of Zhejiang (No. 2015KYA241), the B-Class Project of Enze Group (No. 15EZB2), and the A-Class Project of Taizhou Science and Technology Bureau (No. 14SF07).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shen, Y., Chi, H., Zhou, J. et al. Structure-Based Stepwise Screening of PPARγ Antagonists as Potential Competitors with NCOA1 Coactivator Peptide for PPARγ CIS Site. Int J Pept Res Ther 25, 1369–1377 (2019). https://doi.org/10.1007/s10989-018-9782-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-018-9782-x