Abstract
COVID-19 is an epidemic virus arising from a freshly discovered coronavirus. Most people involved with the coronavirus will experience slight to moderate respiratory disease and recover without needing particular therapy. In this work, the atomic stability of the coronavirus at different thermodynamic properties such as temperature and pressure, was studied. For this purpose, the manner of this virus by atomic precession was described with a molecular dynamics approach. For the atomic stability of coronavirus description, physical properties such as temperature, total energy, volume variation, and atomic force of this structure were reported. In molecular dynamics approach, coronavirus is precisely simulated via S, O, N, and C atoms and performed Dreiding force field to describe these atoms interaction in the virus. Simulation results show that coronavirus stability has reciprocal relation with atomic temperature and pressure. Numerically, after 2.5 ns simulation, the potential energy varies from − 31,163 to − 26,041 eV by temperature changes from 300 to 400 K. Furthermore, this physical parameter decreases to − 28,045 eV rate at 300 K and 2 bar pressure. The volume of coronavirus is another crucial parameter to the stability description of this structure. The simulation shows that coronavirus volume 92% and 14% increases by 100 K and 2 bar variation of simulation temperature and pressure, respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
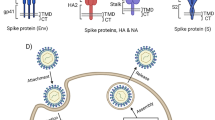
COVID-19 is an epidemic virus arising from a freshly discovered coronavirus. Most people involved with the coronavirus will experience slight to moderate respiratory disease and recover without needing particular therapy. Historically, coronaviruses were reported in the 1930s for the first time when some infection bronchitis virus has appeared from domesticated chickens shown [1] (see Fig. 1). Working at nanoscales levels such as nanoparticles dispersed in the base fluid might help to investigate the virus configuration [2, 3] through the porous medium [4]. The structure of coronaviruses is massive, occasionally pleomorphic (varying in shape), mainly spherical, particles with bulbous surface schemes [5]. The electron micrographs show that the envelope of coronavirus becomes visible as a distinct pair of electron-dense shells (is used to scan the virus particle, shells which are comparatively opaque to the electron beam) [6, 7]. The viral envelope contains a lipid bilayer that it would surround, and spike structural proteins are attached [8]. The proportion of envelope/spike/membrane in the lipid bilayer is about 1:20:300 [9]. Indifferently a coronavirus particle contains 74 surface spikes [10]. A subset of coronaviruses has a shorter spike similar to surface protein (hemagglutinin esterase) [11]. The spikes on the surface of coronavirus are homotrimers of the S protein, which is formed of an S1 and S2 subunit. One of the class I fusion protein is the homotrimeric S protein that intercedes the receptor binding and membrane fusion between the cell and virus that be a host. The head of the spike is the S1 subunit forms, and it consists of the receptor-binding domain. One of the subunit forms (S2) is the stem, which attaches the spike in the viral envelope and on protease activation abilities to fusion. The M and E protein are significant in shaping the viral envelope and preserving its structural shape. In this envelope, there is the nucleocapsid, which is contained from multiple copies of the nucleocapsid (N) protein, bound to the positive-sense single-stranded RNA genome in a going on beads on a string type organization [12]. The preservation of the virus outside the host cell is done by the nucleocapsid, membrane proteins, and lipid bilayer envelope [13]. At this time, there are no particular vaccines or cure for coronavirus.
The typical structure and shape of the coronavirus recognized by experiment researches [37]
Nevertheless, various factors, such as temperature and pressure, appear to affect the manner of this virus and change its stability. Many works have reported the characteristics of this virus [14]. Lu et al. phylogenetic research report that bats might be one of the original hosts of coronavirus. An animal sold in Wuhan might represent an intermediate host, making possible the emergence of the virus in humans [15].
Furthermore, the analysis related to the structure of this research group reported that coronavirus could have the ability to connect to the angiotensin-converting enzyme two receptors in humans [16, 17]. It is not known how the coronavirus behaves in various environmental conditions. In this work, the atomic stability of coronavirus with the molecular dynamics (MD) technique was simulated. MD method is the computer simulations capable of predicting the nuclear manner of a variety of atomic structures [18,19,20]. This calculational approach is broadly used in nanostructures research [21, 22]. In this work, MD simulations were performed to predict the coronavirus atomic stability at various temperatures (T = 300–400 K) and pressures (P = 0–2 bar). For this purpose, physical parameters such as temperature, potential energy, and volume of coronavirus in the aqueous environment were reported.
Methods
In these molecular dynamics simulations, it is used equilibrium MD simulations are used to describe the atomic stability of coronavirus in various temperatures and pressures. The molecular dynamics simulation method is a numerical approach to calculate the dynamical evolution of molecules and atoms. In this process, simulated particles are permitted to interact in the simulation box and give a sight of the dynamic changes of the system. All molecular dynamics simulations in this research were done via employing the Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) provided by Sandia National Laboratories [23,24,25]. To use of this MD simulation package to estimate the atomic stability of coronavirus with various temperatures (T = 300–400 K) and pressures (P = 0–2 bar), the coronavirus molecules were simulated by the atomic accuracy (Fig. 2). This nuclear structure was visualized by Open Visualization Tool (OVITO) [26].
In these MD simulations, aqueous environment is prepared by the Packmol package [27, 28]. Figure 3 shows the simulation box at the front, top, and perspective views. For coronavirus MD simulation in an aqueous environment, the directions x, y, and z were used to show the periodic boundary conditions [29]. To duplicate the canonical phase-space dispersion, the use of this thermostat strives. This was accomplished by altering the equations of motion to contain a non-Newtonian term to preserve the total kinetic energy of the atomic system constant. For the modified equation of atom movement, it can be written:
where ζ is the thermodynamic friction coefficient, defined by Eq. (2):
In this equation, Q is a parameter that clears the time scale of the temperature fluctuation, and X is the number of degrees of freedom. After 2.5 ns MD simulation, the simulated atomic structures were equilibrated at initial condition (initial temperature and pressure). Furthermore, to MD simulate the atomic structure of coronavirus, the Dreiding force field was used [30]. The best choice of this interatomic force field is to affect biological structures. The potential energy for coronavirus atoms in the Dreiding force field was shown as a superposition of bonded and non-bonded ones. Non-bond interaction between atoms in the Dreiding force field is described by the Lennard–Jones (LJ) potential [31]:
Both ε and σ parameters rely on the type of atoms in simulated structures in MD simulations. The energy and cutoff radius and length scale parameter, for different atoms in coronavirus simulation, are written in Table 1.
The bonded interaction in the Dreiding force field consists of bond strength, bond angle bend, and dihedral angle torsion terms. The bond, angle, and dihedral strength stretch in this force field described by harmonic oscillator equations as (4), (5), and (6):
These sympathetic constants in coronavirus MD simulation were selected at 300 (kcal/mol)/A2 and 100 (kcal/mol)/degree2, while d = + 1 or − 1. Furthermore, the equilibrium value of angles and atomic bond length in these MD simulations is reported in Tables 2 and 3.
To aqueous environment definition in this computational work, H2O molecules are simulated by the SPC model. This simple model can describe the atomic evolution of the H2O structure very well in the presence of coronavirus [32]. After atomic modeling and force field implementation, the atomic manner of coronavirus in various temperatures and pressures was simulated. Mathematically, in MD simulation, the location \({r}_{\rm i}\left(t\right)\)and the momentum \({p}_{\rm i}(t)\) are obtained by solving Newton’s second equation of motion,
The momentum \(P_{\rm i}\) can be defined by Eq. (8),
The total energy can be expressed in the form of Hamilton. For N atoms as Eq. (9),
Also, the forces of each atom are related to the potential function as Eq. (10),
Computationally, in MD simulations, the Verlet algorithm is used to find out Newton’s equation of motion as Eqs. (11) and (12),
The average kinetic energy is as Eq. (13),
The instantaneous temperature fluctuates obtained from the Eq. (14),
where \(N_{{{\text{sf}}}}\) is the freedom degree number of the atomic system. The pressure is calculated using the Virial state equation,
The potential force between the atoms \(U\left( {r^{\text{N}}} \right)\) is assumed for each pair of particles. Therefore, for the number of N particles,
This formalism is used in this work for describing the atomic manner (atomic stability) of coronavirus at different temperatures and pressure. The MD simulations are given in two steps:
Step A Coronavirus in the aqueous environment was simulated at 300 K and P = 0 bar with Δt = 1 fs (time step); 150 Å length in x, y, and z directions. In the equilibration process, the NPT ensemble with 0.01 and 1 for temperature and pressure damping rate (respectively) for 2.5 ns was used [33, 34].
Step B The temperature and pressure of simulated structures increase to 400 K and 2 bar after 2.5 ns. For the MD study of coronavirus stability in defined conditions, parameters such as temperature, total energy, and volume of coronavirus were reported.
Results
Equilibration of simulated structures
The atomic structure of coronavirus with H2O molecules in T = 300 K and P = 0 bar is studied, and final position of these structures is used in the second step of this MD study for the first time [35,36,37]. The results showed that the atomic model of coronavirus, which used in this MD study, is adopted with the Dreiding force field. Physically, atomic structures’ stability is described by temperature and potential energy of them at T = 300 K and P = 0 bar. Temperature variation of simulated structures depicted in Fig. 4. From this figure, it can be said that virus and H2O molecules temperature equilibrated after 2.5 ns. Figure 5 displays the potential energy variation of simulated structures in initial condition (temperature and pressure). It can be seen that the atomic structures’ potential energy converged after 2.5 ns. Numerically, the potential power of coronavirus in aqueous environment converged to − 31,163 eV. From the result of this section of this report, it can be said that the Dreiding force field has excellent ability in coronavirus MD simulation.
Atomic stability of coronavirus in various temperature and pressure
For the atomic stability study of coronavirus in various temperature and pressure, first, temperature and potential energy variation in simulated structures were reported. Figure 6 shows that atomic structures’ temperature reaches 350 and 400 K after 2.5 ns. As can be seen, a similar atomic manner for pressure variation of these structures and pressure of them reach after 2.5 ns to 1 bar and 2 bar. By equilibrium of atomic arrangements in the initial condition, the potential energy variation of these atoms was reported. These MD results show that by increasing the temperature of structures, their atomic stability decreases. Numerically, by increasing the temperature of coronavirus in the aqueous environment from 300 to 400 K, the potential energy of them decreases from − 31,163 to − 26,041 eV (Fig. 7 and Table 4). Physically, by increasing the temperature in the MD simulation box, various atoms in coronavirus structure move away and interatomic distance increases in this virus. The distance of atoms in simulated structures has a reciprocal relation with potential energy. So, by increasing the temperature, the potential energy of the system and stability of them decreases.
Furthermore, by increasing the pressure in simulated structures, the distance between atoms decreases. After this atomic phenomenon, repulsive interaction between atoms occurs, which causes an increase in atomic distance between them. By this atomic evolution, the potential energy and atomic stability of coronavirus in the aqueous environment decreases. Numerically by increasing the pressure from 0 to 2 bar, the potential energy of coronavirus in the aqueous environment decreases from − 31,163 to − 28,045 eV (see Fig. 8 and Table 5).
The atomic volume of coronavirus is proportional to its atomic stability in the aqueous environment. These MD results show that by MD simulation time passing, the volume of coronavirus increases (see Fig. 9). Physically, by increasing the simulation temperature and pressure, the distance of atoms in coronavirus structure increases and the stability of this virus decreases. From Fig. 10, by increasing the temperature from 300 to 400 K, the volume of coronaviruses increases from 179,091 to 343,437 Å3. Pressure increasing has a similar effect on virus atomic volume and so on atomic stability. These simulation results show that by increasing the pressure from 0 to 2 bar, the volume of simulated structure changes from 179,091 to 205,283 Å3 (Fig. 11 and Table 6). This atomic behavior indicates that the temperature and pressure increase can be used for this virus disinfection in medical applications. These physical processes can play an effective role in the elimination of coronavirus. Furthermore, from MD simulation results, it can be said that the temperature effect is more effective on atomic disinfection of coronavirus.
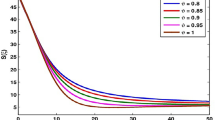
The mutual force between the coronavirus and H2O molecules describes virus evolution in MD simulations. In the final section of this study, the inserted force to coronavirus from the aqueous environment was calculated. Figure 12 shows this physical parameter for virus atoms in T = 300 K after 2.5 ns. The maximum rate of this parameter is 41 eV Å−1. After temperature increasing from 300 to 400 K, the maximum rate of this force reaches 34 eV Å−1 (Table 7). Physically, this parameter decreasing shows the interatomic potential decreasing, too. So it can be said that the atomic stability of the virus decreases by increase in temperature, and the coronavirus can be destroyed in high temperatures. Pressure increasing has a similar effect on the coronavirus atomic manner. By increasing the pressure from 0 to 2 bar, the maximum rate of inserted atomic force to coronavirus decreases from 41 to 37 eV Å−1. Figure 13 shows the inserted force to coronavirus atoms in T = 300 K and P = 2 bar after 2.5 ns.
Conclusions
The present study investigates the atomic stability of coronavirus in an aqueous environment with MD simulations. In these calculations, coronavirus is shown via S, O, N, and C atoms and H2O molecules modeled by SPC. Also, to simulate interatomic force between virus atoms, the Dreiding force field has been selected. The result of these MD simulations shows that the increase in temperature and pressure causes coronavirus disinfection, which can be used for medical purposes. Results are as following:
-
The total energy of this virus in aqueous environment converged to − 31,163 eV after 2.5 ns at T = 300 K and P = 0 bar which implies the atomic stability of simulated structures.
-
The potential energy of coronavirus in the aqueous environment decreases by increase in temperature. This atomic manner shows that coronavirus stability and MD simulation temperature have a reciprocal relation.
-
Numerically, by increasing the temperature from 300 to 400 K, the potential energy of coronavirus in the aqueous environment decreases from − 31,163 to − 26,041 eV.
-
The potential energy of coronavirus in the aqueous environment decreases by increasing the pressure. So atomic stability of these structures has reciprocal relation with MD simulation pressure.
-
Numerically, by increasing the pressure from 0 to 2 bar, the potential energy of coronavirus in the aqueous environment decreases from − 31,163 to − 28,045 eV.
-
The volume of the coronavirus increases by increase in temperature in the MD simulation box. This atomic manner shows that unstability was growing in the virus by increase in temperature. Numerically, coronavirus volume increases from 179,091 to 343,437 Å3 after temperature increasing from 300 to 400 K.
-
The volume of the coronavirus in the aqueous environment increases by increase in pressure in the MD simulation box. Numerically, coronavirus volume increases from 179,091 to 205,283 Å3 after pressure increases from 0 to 1 bar.
-
Coronavirus and aqueous environment mutual atomic force changes from 41 to 34 eV Å−1 by the change in thermodynamic properties such as temperature and pressure variations.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Fan Y, Zhao K, Shi ZL, Zhou P. Bat coronaviruses in China. Viruses. 2019;11(3):210.
Saif RS, Muhammad T, Sadia H, Ellahi R. Hydromagnetic flow of Jeffrey nanofluid due to a curved stretching surface. Phys A Stat Mech Appl. 2020;551:124060.
Riaz A, Zeeshan A, Bhatti MM, Ellahi R. Peristaltic propulsion of Jeffrey nano-liquid and heat transfer through a symmetrical duct with moving walls in a porous medium. Phys A. 2020;545:123788.
Alamri SZ, Ellahi R, Shehzad N, Zeeshan A. Convective radiative plane Poiseuille flow of nanofluid through porous medium with slip: an application of Stefan blowing. J Mol Liq. 2019;273:292–304.
Goldsmith CS, Tatti KM, Ksiazek TG, Rollin PE, Comer JA, Lee WW, et al. Ultrastructural characterization of SARS coronavirus. Emerg Infect Dis. 2004;10(2):320–6.
Neuman BW, Adair BD, Yoshioka C, Quispe JD, Orca G, Kuhn P, et al. Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy. J Virol. 2006;80(16):7918–28.
Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. In: Maier HJ, Bickerton E, Britton P, editors. Coronaviruses methods in molecular biology, vol. 1282. Berlin: Springer; 2015. p. 1–23.
Lai MM, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100.
Cavanagh D, Mawditt K, Sharma M, Drury SE, Ainsworth HL, Britton P, Gough RE. Detection of a coronavirus from turkey poults in Europe genetically related to infectious bronchitis virus of chickens. In: Schmidt A, Weber O, Wolff MH, editors. Avian pathology Birkhäuser advances in infectious diseases BAID, vol 30. Basel: Birkhäuser; 2001. p. 355–68.
Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh M, Connelly S, Droese B, Klaus JP, Makino S, Sawicki SG, Siddell SG. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22.
de Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, Perlman S, Poon L, Rottier PJ, Talbot PJ, Woo PC, Ziebuhr J. Family coronaviridae. In: King AM, Lefkowitz E, Adams MJ, Carstens EB, editors. International committee on taxonomy of viruses, international union of microbiological societies. Virology Division. Ninth report of the international committee on taxonomy of viruses. Oxford: Elsevier; 2011. p. 806–28.
Chang CK, Hou MH, Chang CF, Hsiao CD, Huang TH. The SARS coronavirus nucleocapsid protein—forms and functions. Antiviral Res. 2014;103:39–50.
Neuman BW, Kiss G, Kunding AH, Bhella D, Baksh MF, Connelly S, et al. A structural analysis of M protein in coronavirus assembly and morphology. J Struct Biol. 2011;174(1):11–22.
Masters PS. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207.
Lu R, Sca M, Zhao X, Da M, Li J. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;327(10224):565–74.
Cui J, Li F, Shi Z-L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2018;17:181–92.
Alder BJ, Wainwright TE. Studies in molecular dynamics. I General method J Chem Phys. 1959;31(2):459.
Rahman A. Correlations in the motion of atoms in liquid argon. Phys Rev. 1964;136(2A):A405–11.
Sabetvand R, Ghazi ME, Izadifard M. Studying temperature effects on electronic and optical properties of cubic CH3NH3SnI3 perovskite. J Comput Electron. 2020;19:70–9.
Ghanbari A, Warchomicka F, Sommitsch C, Zamanian A. Investigation of the oxidation mechanism of dopamine functionalization in an AZ31 magnesium alloy for biomedical applications. Coatings. 2019;9(9):584.
Jolfaei NA, Jolfaei NA, Hekmatifar M, Piranfar A, Toghraie D, Sabetvand R, Rostami S. Investigation of thermal properties of DNA structure with precise atomic arrangement via equilibrium and non-equilibrium molecular dynamics approaches. Comput Methods Programs Biomed. 2019;185:105169.
Ashkezari AZ, Jolfaei NA, Jolfaei NA, Hekmatifar M, Toghraie D, Sabetvand R, Rostami S. Calculation of the thermal conductivity of human serum albumin (HSA) with equilibrium/non-equilibrium molecular dynamics approaches. Comput Methods Programs Biomed. 2020;188:105256.
Brown WM, Wang P, Plimpton SJ, Tharrington AN. Implementing molecular dynamics on hybrid high performance computers—short range forces. Comput Phys Commun. 2011;182:898–911.
Brown WM, Kohlmeyer A, Plimpton SJ, Tharrington AN. Implementing molecular dynamics on hybrid high performance computers—particle–particle particle-mesh. Comput Phys Commun. 2012;183(3):449–59.
Martinez L, Andrade R, Birgin EG, Martínez JM. PACKMOL: a package for building initial configurations for molecular dynamics simulations. J Comput Chem. 2009;30(13):2157–64.
Stukowski A. Visualization and analysis of atomistic simulation data with OVITO—the open visualization tool. Model Simul Mater Sci Eng. 2009;18(1):015012.
Martínez L, Andrade R, Birgin EG, Martínez JM. PACKMOL: a package for building initial configurations for molecular dynamics simulations. J Comput Chem. 2009;30(13):2157–64.
Martínez JM, Martínez L. Packing optimization for automated generation of complex system’s initial configurations for molecular dynamics and docking. J Comput Chem. 2003;24(7):819–25.
Rapaport DC. The art of molecular dynamics simulation (2nd ed.). Cambridge: Cambridge University Press. ISBN 0-521-82568-7. 2004
Mayo SL, Olafson BD, Goddard WA. DREIDING: a generic force-field for molecular simulations. J Phys Chem. 1990;94(26):8897–909.
Lennard-Jones JE. On the determination of molecular fields. Proc R Soc Lond A. 1924;106(738):463–77.
Berendsen HJC, Postma JPM, van Gunsteren WF, Hermans J. In: B. Pullmanm, editor. Intermolecular forces. Dordrecht: Reidel, 1981.
Nosé S. A unified formulation of the constant temperature molecular-dynamics methods. J Chem Phys. 1984;81(1):511–9.
Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31(3):1695–7.
Qi C, Tang J, Wang G. Natural convection of composite nanofluids based on a two-phase lattice Boltzmann model. J Therm Anal Calorim. 2020. https://doi.org/10.1007/s10973-020-09519-9.
Qi C, Wang G, Yang L, Wan Y, Rao Z. Two-phase lattice Boltzmann simulation of the effects of base fluid and nanoparticle size on natural convection heat transfer of nanofluid. Int J Heat Mass Transf. 2017;105:664–72.
Almeida JD, Berry DM, Cunningham CH, Hamre D, Hofstad MS, Mallucci L, McIntosh K, Tyrrell DA. Virology: coronaviruses. Nature. 1968;220(5168):650. https://doi.org/10.1038/220650b0.
Funding
This research received no external funding.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Malekahmadi, O., Zarei, A., Botlani Esfahani, M.B. et al. Thermal and hydrodynamic properties of coronavirus at various temperature and pressure via molecular dynamics approach. J Therm Anal Calorim 143, 2841–2850 (2021). https://doi.org/10.1007/s10973-020-10353-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10973-020-10353-2