Abstract
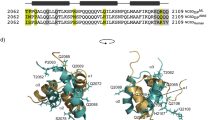
Protein folding thermodynamics can determine the contribution of protein fluctuation, which is difficult to detect but critical for protein function. We have analyzed the structural dynamic properties of the DNA-binding domain of a transcriptional activator, c-Myb R2R3. Earlier reports state that a substitution at the linker between R2 and R3 resulted in a significant loss of affinity toward the cognate DNA, mainly due to increased entropy in the DNA-unbound state. In this study, we analyzed the effects of the linker on folding stability and found that mutation of Pro140 to Gly or Ala resulted in decreased stability, due to an unfavorable enthalpy change that was only partially compensated by a favorable entropy change. Considering that the mutation would increase protein fluctuations in both the folded and unfolded states, we assume that the change in the folded state would be larger than in the unfolded state. This is perhaps due to the additional intramolecular interactions that only occur in the folded state. The increase in protein fluctuation in the folded state post-mutation was correlated with protein function, as reported previously.



Similar content being viewed by others
References
Naganathan AN, Doshi U, Fung A, Sadqi M, Muñoz V. Dynamics, energetics, and structure in protein folding. Biochemistry. 2006;45:8466–75.
Wand AJ. The dark energy of proteins comes to light: conformational entropy and its role in protein function revealed by NMR relaxation. Curr Opin Struct Biol. 2013;23:75–81.
Biedenkapp H, Borgmeyer U, Sippel AE, Klempnauer KH. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature. 1988;335:835–7.
Sakura H, Kanei-Ishii C, Nagase T, Nakagoshi H, Gonda TJ, Ishii S. Delineation of three functional domains of the transcriptional activator encoded by the c-myb protooncogene. Proc Natl Acad Sci USA. 1989;86:5758–62.
Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–48.
Ogata K, Morikawa S, Nakamura H, Hojo H, Yoshimura S, Zhang R, Aimoto S, Ametani Y, Hirata Z, Sarai A, Ishii S, Nishimura Y. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nature Struct Biol. 1995;2:309–20.
Oda M, Furukawa K, Ogata K, Sarai A, Ishii S, Nishimura Y, Nakamura H. Investigation of the pyrimidine preference by the c-Myb DNA-binding domain at the initial base of the consensus sequence. J Biol Chem. 1997;272:17966–71.
Oda M, Furukawa K, Ogata K, Sarai A, Ishii S, Nishimura Y, Nakamura H. Identification of indispensable residues for specific DNA-binding in the imperfect tandem repeats of c-Myb R2R3. Protein Eng. 1997;10:1407–14.
Oda M, Furukawa K, Sarai A, Nakamura H. Kinetic analysis of DNA binding by the c-Myb DNA-binding domain using surface plasmon resonance. FEBS Lett. 1999;454:288–92.
Morii H, Uedaira H, Ogata K, Ishii S, Sarai A. Shape and energetics of a cavity in c-Myb probed by natural and non-natural amino acid mutations. J Mol Biol. 1999;292:909–20.
Inaba S, Fukada H, Ikegami T, Oda M. Thermodynamic effects of multiple protein conformations on stability and DNA binding. Arch Biochem Biophys. 2013;537:225–32.
Oda M, Furukawa K, Ogata K, Sarai A, Nakamura H. Thermodynamics of specific and non-specific DNA binding by the c-Myb DNA-binding domain. J Mol Biol. 1998;276:571–90.
Spolar RS, Record MT Jr. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–84.
Dunitz JD. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chem Biol. 1995;2:709–12.
Matthews BW, Nicholson H, Becktel WJ. Enhanced protein thermostability from site-directed mutations that decrease the entropy of unfolding. Proc Natl Acad Sci USA. 1987;84:6663–7.
Yutani K, Hayashi S, Sugisaki Y, Ogasahara K. Role of conserved proline residues in stabilizing tryptophan synthase subunit: analysis by mutants with alanine or glycine. Proteins Struct Funct Genet. 1991;9:90–8.
Inaba S, Maeno A, Sakurai K, Puthenpurackal SN, Ikegami T, Akasaka K, Oda M Molecular strategy of c-Myb DNA-binding domain for function: Low-populated unfolding species revealed from temperature-dependent studies. under review.
Dyson HJ. Roles of intrinsic disorder in protein–nucleic acid interactions. Mol BioSyst. 2012;8:97–104.
Chu H-L, Chen T-H, Wu C-Y, Yang Y-C, Tseng S-H, Cheng T-M, Ho L-P, Tsai L-Y, Li H-Y, Chang C-S, Chang C-C. Thermal stability and folding kinetics analysis of disordered protein, securing. J Therm Anal Calorim. 2014;115:2171–8.
Oda M, Nakamura H. Thermodynamic and kinetic analyses for understanding sequence-specific DNA recognition. Genes Cells. 2000;5:319–26.
Ogata K, Kanei-Ishii C, Sasaki M, Hatanaka H, Nagadoi A, Enari M, Nakamura H, Nishimura Y, Ishii S, Sarai A. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol. 1996;3:178–87.
Puthenpurackal SN, Maeno A, Matsuo H, Oda M, Morii H, Akasaka K. Extensively hydrated but folded: a novel state of globular proteins stabilized at high pressure and low temperature. Biophys J. 2012;102:L08–10.
Author information
Authors and Affiliations
Corresponding author
Additional information
The present article is based on the lecture presented at JCCTA50 conference in Osaka – Japan on 28–30 September, 2014.
Rights and permissions
About this article
Cite this article
Inaba, S., Fukada, H. & Oda, M. Thermodynamic effects of a linker region between two repeats of a protein, c-Myb R2R3, on its stability and structural dynamics. J Therm Anal Calorim 123, 1763–1767 (2016). https://doi.org/10.1007/s10973-015-4812-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10973-015-4812-9