Abstract
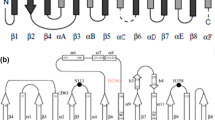
Recent investigations of Aneurinibacillus thermoaerophilus strains have allowed identification of a unique solvent tolerant lipase, distinct from known lipases. This work reports the expression and purification of this lipase (LipAT) and the first characterization of its structure and temperature and pH-dependent behaviour. LipAT has a secondary structural content compatible with the canonical lipase α/β hydrolase fold, and is dimeric at neutral pH. The protein was folded from pH 5 to 10, and association into folded aggregates at pH 7 and 8 likely protected its secondary structures from thermal unfolding. The enzyme was active from 25 to 65 °C under neutral pH, but its maximal activity was detected at pH 10 and 45 °C. The ability of LipAT to recover from high temperature was investigated. Heating at 70 °C and pH 10 followed by cooling prevented the restoration of activity, while similar treatments performed at pH 8 (where folded aggregates may form) allowed recovery of 50% of the initial activity. In silico analyses revealed a high conservation (85% or more) for the main lipase signature sequences in LipAT despite an overall low residue identity (60% identity compared to family I.5 lipases). In contrast, the active site lid region in LipAT is very distinct showing only 25% amino acid sequence identity to other homologous lipases in this region. Comparison of lids among lipases from the I.5 family members and LipAT reveals that this region should be a primary target for elucidation, optimisation and prediction of structure–function relationships in lipases.







Similar content being viewed by others
References
Wakhlu J (2006) Yeast lipases: enzyme purification, biochemical properties and gene cloning. Electron J Biotechnol 9(1):0717–3458
Gandhi N (1997) Applications of lipases. JAOCS 74(6):621–634
Secundo F, Carrea G, Tarabiono C, Gatti-Lafranconi P, Brocca S, Lotti M, Jaeger KE, Puls M, Eggert T (2006) The lid is a structural and functional determinant of lipase activity and selectivity. J Mol Catal B 39(1–4):166–170
Global markets for enzymes in industrial applications (BIO030G). BCC Research. June 2014, visited June 7th 2016. http://www.bccresearch.com/market-research/biotechnology/enzymes-industrial-applications-bio030h.html
Gupta R, Gupta N, Rathi P (2007) Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol 64(6):763–781
Fujinami S, Fujisawa M (2010) Industrial applications of alkaliphiles and their enzymes past, present and future. Environ Technol 31(8–9):845–856
Charbonneau DM, Meddeb-Mouelhi F, Beauregard M (2010) A novel thermostable carboxylesterase from Geobacillus thermodenitrificans: evidence for a new carboxylesterase family. J Biochem 148(3):299–308
Rahman RNZRA, Masomian M, Salleh AB, Basri M (2009) A new thermostable lipase by Aneurinibacillus thermoaerophilus strain HZ: nutritional studies. Ann Microbiol 59(1):133–139
Masomian M, Rahman RNZRA, Salleh AB, Basri M (2010) A unique thermostable and organic solvent tolerant lipase from newly isolated Aneurinibacillus thermoaerophilus strain HZ: physical factor studies. World J Microbiol Biotechnol 26(9):1693–1701
Masomian M, Rahman RNZRA, Salleh AB, Basri M (2016) Analysis of a Comparative Sequence and genomic Data to Verify Phylogenetic Relationship and Explore a New Subfamily of Bacterial lipase. PLoS ONE 11(3):e0149851
Bradford M (1976) A dye binding assay for protein. Anal Biochem 72:248–254
Kouker G, Jaeger K-E (1986) Specific and sensitive plate assay for bacterial lipases. Appl Environ Microbiol 53(1):211–213
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TM (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5-W9
Chakravorty D, Parameswaran S, Kumar Dubey V, Patra S (2011) In silico characterization of thermostable lipases. Extremophiles 15(1):89–103
Pettersen E, Goddard T, Huang C, Couch G, Greenblatt D, Meng E, Ferrin T (2004) UCSF Chimera—a visualization system for exploratory research and analysis. InterScience 25(13):1605–1612
Rahman RNZRA, Leow T, Salleh AB, Basri M (2007) Geobacillus zalihae sp. novel thermophilic lipolytic bacterium isolated from palm oil mill effluent in Malaysia. BMC Microbiol 7(1):77
Hooft R W W, Vriend G, Sander C, Abola EE (1996) Errors in protein structures. Nature 381(6580):272
Laskowski RA, MacArthur MW, Moss D, Thornton (1993) J M, PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291
Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22(2):195–201
Johnson WC (1999) Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 35(3):307–312
Sreerama N, Woody RW (2000) Estimation of protein secondary structures from circular dichroism spectra: comparison of CONTIN, SELCON and CDSSTR with an expended reference set. Anal Biochem 287(2):252–260
Charbonneau DM, Beauregard M (2013) Role of key salt bridges in thermostability of G. thermodenitrificans EstGtA2: distinctive patterns within the new bacterial lipolytic enzyme family XV. PLoS ONE 8(10):e76675
Pace CN, Shirley BA, Thompson JA (1989) Measuring the conformational stability of a protein. In:Creighton TE (ed) Protein structure-a practical approach. Oxford University Press, Oxford, pp 311–330
Zottig X, Meddeb-Mouelhi F, Beauregard M (2016) Development of a high throughput liquid state assay for lipase activity using natural substrates and rhodamine B. Anal Biochem 496:25–29
Timucin E, Sezerman OU The conserved lid tryptophan, W211, potentiates thermostability and thermoactivity in bacterial thermoalkalophilic lipases. PLoS ONE 8(12):e85186. doi:10.1371/journal.pone.0085186
Li H, Zhang X (2005) Characterization of thermostable lipase from thermophilic Geobacillus sp. TW1. Protein Expr Purif 42(1):153–159
Balan A, Ibrahim D, Rahim Abdul, R and Azzahra Ahmad Rashid F (2012) Purification and characterization of a thermostable lipase from Geobacillus thermodenitrificans IBRL-nra. Enzyme Res. doi:10.1155/2012/987523
Korman TP, Sahachartsiri B, Charbonneau DM, Huang GL, Beauregard M, Bowie JU (2013) Dieselzymes: development of a stable and methanol tolerant lipase for biodiesel production by directed evolution. Biotechnol Biofuels 6:70–83
Arpigny JL, Jaeger KE (1999) Bacterial lipolytic enzymes classification and properties. Biochem J 343(1):177–183
Fisher M, Pleiss (1998) The lipase engineering database: a navigation and analysis tool for proteins families. Nucleic Acids Res 31(1):319–321
Brocca S, Secundo F, Ossola M, Alberghina L, Carrea G, Lotti M (2003) Sequence of the lid affects activity and specificity of Candida rugose lipase isoenzymes. Protein Sci 12(10):2312–2319
Xiao X, Lowe ME (2015) The β5-Loop and Lid Domain Contribute to the Substrate Specificity of Pancreatic Lipase-related Protein 2 (PNLIPRP2). J Biol Chem 290(48):28847–28856. doi: 10.1074/jbc.M115.683375
Yu XW, Tan NJ, Xiao R, Xu Y (2012) Engineering a disulfide bond in the lid hinge region of Rhizopus chinensis lipase: increased thermostability and altered acyl chain length specificity. PLoS ONE 7(10):e46388. doi: 10.1371/journal.pone.0046388
Acknowledgements
X.Z. acknowledges a scholarship from CRMR (U. Laval, Canada). The editorial help of Pr. J. Turnbull in preparing this manuscript is acknowledged.
Funding
This work was supported by a NSERC Discovery grant awarded to M.B.
Author information
Authors and Affiliations
Contributions
XZ carried out all experimental work and wrote the initial draft. FM collaborated to production and purification protocols, and performed editorial work. DC collaborated to the in silico analyses and performed editorial work. MB supervised the research and wrote the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zottig, X., Meddeb-Mouelhi, F., Charbonneau, D.M. et al. Characterization of a Novel Alkalophilic Lipase From Aneurinibacillus thermoaerophilus: Lid Heterogeneity and Assignment to Family I.5. Protein J 36, 478–488 (2017). https://doi.org/10.1007/s10930-017-9743-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10930-017-9743-9