Abstract
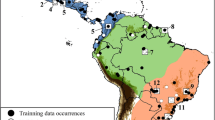
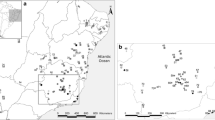
The white-eared opossum, Didelphis albiventris Lund, 1840, is one of the most widely distributed South American marsupials. As currently defined, the species ranges from northeastern and central Brazil through Paraguay and central Bolivia, all of Uruguay, and in Argentina, from the north to northern Patagonia. Previous phylogeographic analyses found two haplogroups, one corresponding to northeastern and central Brazil and the other to southern Brazil, Bolivia, and Paraguay. In those studies, differentiation was based on the geographic distance between both regions and the presence of geographic barriers among populations. The aims of the present contribution were to perform population-genetic analyses using the mitochondrial markers cytochrome c oxidase 1 (COI) and cytochrome b (cyt b) in order to elucidate the phylogeographic history of this species by incorporating samples from Argentina into the published datasets, and to evaluate whether there is more than one taxonomic entity under the name Didelphis albiventris. We studied the craniodental morphology of the species and used environmental niche modeling (ENM) as an additional methodology to investigate the environmental factors affecting its distribution. Our molecular, morphological, and ENM results strongly suggest the existence of two taxonomic entities under Didelphis albiventris, one in southern Brazil, Argentina, Paraguay, Uruguay, and Bolivia, and the other in central and northern Brazil. Data from specimens in the missing middle part of the taxon’s range and evidence from nuclear markers are needed before formalizing a taxonomic split.








Similar content being viewed by others
References
Anderson S (1997) Mammals of Bolivia, taxonomy and distribution. Bull Amer Mus Nat Hist 231:1–652
Astúa D (2015) Order Didelphimorphia. In: Wilson DE, Mittermeier RE (eds) Handbook of the Mammals of the World, Vol. 5 Monotremes and Marsupials. Lynx Editions, Barcelona, pp 70–187
Barve N, Barve V, Jiménez-Valverde A, Lira-Noriega A, Maher SP, Peterson AT, Soberón J, Villalobos F (2011) The crucial role of the accessible area in ecological niche modeling and species distribution modeling. Ecol Model 222:1810–1819. https://doi.org/10.1016/j.ecolmodel.2011.02.011
Bianchini M (2018) Comadreja overa (Didelphis albiventris): ampliación de su distribución geográfica en su extremo austral (Patagonia) e información de su antigüedad, y aportes a su límite occidental en la Argentina. Not Faun (Segunda Ser) 238:1–13
Bonvicino CR, Lazar A, Freitas TPTd, Lanes RdO, D’Andrea PS (2022) Diversification of South American Didelphid Marsupials. In: Cáceres NC, Dickman CR (eds) American and Australasian Marsupials. Springer, Cham, pp 1–35
Carrera M, Udrizar Sauthier DE (2014) Enlarging the knowledge on Didelphis albiventris (Didelphimorphia: Didelphidae) in northern Patagonia: New records and distribution extension. Hist Nat (Tercera Ser) 4:111–115
Cerqueira R (1980) A study of neotropical Didelphis (Mammalia, Polyprotodontia, Didelphidae). Dissertation, University of London
Cerqueira R (1985) The Distribution of Didelphis in South America (Polyprotodontia, Didelphidae). J Biogeogr 12:135–145
Cerqueira R, Tribe CJ (2008) Genus Didelphis. In: Gardner AL (ed) Mammals of South America Vol. 1. Marsupials, Xenarthrans, Shrews, and Bats. Chicago University Press, Chicago pp 17–25. https://doi.org/10.7208/chicago/9780226282428.001.0001
Chambers JM, Cleveland WS, Kleiner B, Tukey PA (1983) Graphical Methods for Data Analysis. Wadsworth & Brooks, Cole Publishing Company, Pacific Grove
Chemisquy MA, Prevosti FJ, Martínez P, Raimondi V, Cabello Stom JE, Acosta-Jamett G, Montoya-Burgos JI (2019) How many species of grey foxes (Canidae, Carnivora) are in southern South America? Mastozool Neotropical 26:81–91. https://doi.org/10.31687/saremMN.19.26.1.0.16
Chemisquy MA, Morinigo FM, Fameli A, González-Ittig RE (2021) Genetic diversity of the white-eared opossum Didelphis albiventris (Didelphimorphia: Didelphidae) in Argentina. Mastozool Neotropical 28:e0534. https://doi.org/10.31687/saremMN.21.28.1.0.20
Costa LP, Astúa D, Brito D, Soriano P, Lew D (2021) Didelphis albiventris (amended version of 2015 assessment). The IUCN Red List of Threatened Species 2021: e.T40489A197310863. https://doi.org/10.2305/IUCN.UK.2021-1.RLTS.T40489A197310863.en
Costa-Araújo R, Silva-Jr. JS, Boubli JP, Rossi RV, Canale GR, Melo FR, Bertuol F, Silva FE, Silva DA, Nash SD, Sampaio I, Farias IP, Hrbek T (2021) An integrative analysis uncovers a new, pseudo-cryptic species of Amazonian marmoset (Primates: Callitrichidae: Mico) from the arc of deforestation. Sci Rep 11:15665. https://doi.org/10.1038/s41598-021-93943-w
Cozzuol MA, Goin F, de Los Reyes M, Ranzi A (2006) The oldest species of Didelphis (Mammalia, Marsupialia, Didelphidae), from the Late Miocene of Amazonia. J Mamm 87:663–667
Daac O (2017) MODIS and VIIRS Land Products Global Subsetting and Visualization Tool. ORNL DAAC. https://daac.ornl.gov/cgi-bin/dsviewer.pl?ds_id=1379. https://doi.org/10.3334/ORNLDAAC/1379
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772. https://doi.org/10.1038/nmeth.2109
Dayrat B (2005) Towards integrative taxonomy. Biol J Linn Soc 85:407–417. https://doi.org/10.1111/j.1095-8312.2005.00503.x
Dellicour S, Flot J-F (2015) Delimiting species-poor data sets using single molecular markers: a study of barcode gaps, haplowebs and GMYC. Syst Biol 64:900–908. https://doi.org/10.1093/sysbio/syu130
Dias CAR, Perini FA (2018) Biogeography and early emergence of the genus Didelphis (Didelphimorphia, Mammalia). Zool Scr 47:645–654. https://doi.org/10.1111/zsc.12306
Dinerstein E, Olson D, Joshi A et al (2017) An ecoregion-based approach to protecting half the terrestrial realm. BioScience 67:534–545. https://doi.org/10.1093/biosci/bix014
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012). Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29:1969–1973
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molec Ecol Resour 10:564–567. https://doi.org/10.1111/j.1755-0998.2010.02847.x
Ezard T, Fujisawa T, Barraclough TG (2009) SPLITS: SPecies’ LImits by Threshold Statistics. R package version 1.0–18/r45. http://R-Forge.R-project.org/projects/splits/
Fick SE, Hijmans RJ (2017) WorldClim 2: New 1-Km Spatial Resolution Climate Surfaces for Global Land Areas. Int J Climatol 37:4302–4315. https://doi.org/10.1002/joc.5086
Flores DA (2006) Orden Didelphimorphia. In: Barquez RM, Díaz M, Ojeda RA (eds) Mamíferos de Argentina, Sistemática y Distribución. SAREM, San Miguel de Tucumán pp 31–45
Folguera A, Orts D, Spagnuolo M, Rojas Vera E, Litvak V, Sagripanti L, Ramos ME, Ramos VA (2011) A review of Late Cretaceous to Quaternary palaeogeography of the southern Andes. Biol J Linnean Soc 103:250–268. https://doi.org/10.1111/j.1095-8312.2011.01687.x
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3:294–9
Freeman JB, Dale R (2013) Assessing bimodality to detect the presence of a dual cognitive process. Behav Res Methods 45:83–97. https://doi.org/10.3758/s13428-012-0225-x
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147:915–925. https://doi.org/10.1093/genetics/147.2.915
Funk DJ, Nosil P, Etges WJ (2006) Ecological divergence exhibits consistently positive associations with reproductive isolation across disparate taxa. Proc Natl Acad Sci USA 103:3209–3213. https://doi.org/10.1073/pnas.0508653103.
Goin FJ, Candela, AM (2004) New Paleogene marsupials from the Amazon Basin of eastern Perú. In: Campbell Jr KE (ed) The Paleogene Mammalian Fauna of Santa Rosa, Amazonian Peru. Natural History Museum of Los Angeles County, Science Series, Number 40; pp 15–60
Goin FJ, Woodburne MO, Zimicz N, Martin GM, Chornogubsky L (2016) A Brief History of South American Metatherians: Evolutionary Contexts and Intercontinental Dispersals. Springer Netherlands, Amsterdam
González B, Brook F, Martin GM (2022) Updated distribution and conservation perspectives of marmosine opossums from Colombia. Hystrix 33:41–50
Grant WS (2015) Problems and cautions with sequence mismatch analysis and Bayesian skyline plots to infer historical demography. J Heredity 106:333–346
Guillot G, Mortier F, Estoup A (2005) GENELAND: a computer package for landscape genetics. Molec Ecol Notes 5:712–715. https://doi.org/10.1111/j.1471-8286.2005.01031.x
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Hoorn C, Wesselingh FP, Ter Steege H et al. (2010) Amazonia through time: Andean uplift, climate change, landscape evolution and biodiversity. Science 330:927–931. https://doi.org/10.1126/science.1194585
Jayat JP, Teta P, Ojeda AA, Steppan SJ, Osland JM, Ortiz PE, Novillo A, Lanzone C, Ojeda RA (2021) The Phyllotis xanthopygus complex (Rodentia, Cricetidae) in central Andes, systematics and description of a new species. Zool Scr 50:689–706. https://doi.org/10.1111/zsc.12510
Kass JM, Muscarella R, Galante PJ, Bohl CL, Pinilla-Buitrago GE, Boria RA, Soley-Guardia M, Anderson RP (2021) ENMeval 2.0: Redesigned for customizable and reproducible modeling of species’ niches and distributions. Methods Ecol Evol 12:1602–1608. https://doi.org/10.1111/2041-210X.13628
Leigh JW, Bryant D (2015) PopART: Full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116. https://doi.org/10.1111/2041-210x.12410
Lemos B, Cerqueira R (2002) Morphological and morphometric differentiation in the white-eared opossum group (Didelphidae, Didelphis). J Mamm 83:354–69
Luckett P W (1993) An ontogenetic assessment of dental homologies in therian mammals. In: Szalay FS, Novacek MJ, McKenna MC (eds) Mammal Phylogeny: Mesozoic Differentiation, Multituberculates, Monotremes, Early Therians and Marsupials. Springer-Verlag, New York, pp 182–204
Maechler M (2021) diptest: Hartigan’s Dip Test Statistic for Unimodality – Corrected version 0.76. https://cran.r-project.org/
Massoia E, Lartigau B (1995) Mamíferos (Rodentia, Lagomorpha y Marsupicarnivora) cazados por Tyto alba en el Río Limay, Departamento Pilcanuyeu, Provincia de Río Negro. Bol Cient APRONA (IX) 27:15–18
Massoia E, Forasiepi A, Teta P (2000) Los Marsupiales de la Argentina. Editorial L.O.L.A. (Literature of Latin America), Buenos Aires
Merow C, Smith MJ, Silander JA (2013) A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography36:1058–1069.
Miller S, Diker D, Polesky H (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acid Res 6:1215. https://doi.org/10.1093/nar/16.3.1215
Morello J, Matteucci SD, Rodriguez AF, Silva ME (2012) Ecorregiones y Complejos Ecosistémicos de Argentina. 1ra ed. Orientación Gráfica Editora, Buenos Aires
Nascimento DC, Campos BATP, Fraga EC, Barros MC (2019) Genetic variability of populations of the white-eared opossum, Didelphis albiventris Lund 1840 (Didelphimorphia; Didelphidae) in Brazil. Braz J Biol 79:594–602. https://doi.org/10.1590/1519-6984.184842
Padial JM, Miralles A, De la Riva I, Vences M (2010) The integrative future of taxonomy. Front Zool 7:16. https://doi.org/10.1186/1742-9994-7-16
Pante E, Schoelinck C, Puillandre N (2015) From integrative taxonomy to species description: one step beyond. Syst Biol 64:152–160. https://doi.org/10.1093/sysbio/syu083
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach, Bioinformatics 26:419–420. https://doi.org/10.1093/bioinformatics/btp696
Paradis E, Schliep K (2019) ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528
Pastrán-López OGA, Rivero-Castro AG, Ruiz-Estebes EG, Amoni-Sacchi HJ, Sanchez-Castro L (2022) Ampliación de distribución de Didelphis albiventris (Didelphimorphia, Didelphidae) en su extremo austral, provincia de Rio Negro, Argentina. Mamm Notes 8:230. https://doi.org/10.47603/mano.v8n1.230
Phillips SJ, Anderson RP, Dudík M, Schapire RE, Blair ME (2017) Opening the black box: an open-source release of Maxent. Ecography 40:887–893
Pons J, Barraclough TG, Gomez-Zurita J, Cardoso A, Duran D, Hazell S, Kamoun S, Sumlin W, Vogler AP (2006) Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Syst Biol 55:595–609
Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol 23:564–571. https://doi.org/10.1016/j.tree.2008.06.010
QGIS Development Team (2022) QGIS Geographic Information System. Open Source Geospatial Foundation Project.
Qi J, Chehbouni A, Huete AR, Kerr YH, Sorooshian A (1994) A modified soil adjusted vegetation index. Remote Sens Environ 48:119–126
Radosavljevic A, Anderson RP (2014) Making better Maxent models of species distributions: complexity, overfitting and evaluation. J Biogeogr 41:629–643. https://doi.org/10.1111/jbi.12227
Rambaut A (2012) FigTree v1.4. http://tree.bio.ed.ac.uk/software/figtree/
Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904.
Ramos-Onsins S, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19:2092–2100. https://doi.org/10.1093/oxfordjournals.molbev.a004034
Rocha RG, Ferreira E, Loss AC, Heller R, Fonseca C, Pires Costa L (2015) The Araguaia River as an important biogeographical divide for didelphid marsupials in Central Brazil. J Hered 106:593–607. https://doi.org/10.1093/jhered/esv058
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9:552–69. https://doi.org/10.1093/oxfordjournals.molbev.a040727
Salomon N (2011) Evolutionary biogeography and speciation: essay on a synthesis. J Biogeogr 28:13–27. https://doi.org/10.1046/j.1365-2699.2001.00515.x
Schnase JL, Carroll ML, Gill RL, Tamkin GS, Li J, Strong SL, Maxwell TP, Aronne ME, Spradlin CS (2021) Toward a Monte Carlo approach to selecting climate variables in MaxEnt. PLoS ONE 16:e0237208. https://doi.org/10.1371/journal.pone.0237208
Silvestro D, Michalak I (2012) raxmlGUI: a graphical front-end for RAxML. Org Divers Evol 12:335–337. https://doi.org/10.1007/s13127-011-0056-0
Singh BN (2012) Concepts of species and models of speciation. Curr Sci 103:784–790.
Smith P (2009) FAUNA Paraguay Handbook of the Mammals of Paraguay, Volume 1: Marsupialia: Didelphis albiventris. http//www.faunaparaguay.com. Accessed 15 February 2022
Solari S, Sotero-Caio CG, Baker RJ (2019) Advances in systematics of bats: towards a consensus on species delimitation and classifications through integrative taxonomy. J Mamm 100:838–851. https://doi.org/10.1093/jmammal/gyy168
Sousa LCC, Gontijo CMF, Botelho HA, Fonseca CC (2012) Mitochondrial genetic variability of Didelphis albiventris (Didelphimorphia, Didelphidae) in Brazilian localities. Genet Mol Biol 35:522–529. https://doi.org/10.1590/s1415-47572012005000035
Spitzer M, Wildenhain J, Rappsilber J, Tyers M (2014) BoxPlotR: a web tool for generation of box plots. Nat Methods 11:121–122
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. https://doi.org/10.1093/bioinformatics/btl446
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595. https://doi.org/10.1093/genetics/123.3.585
Turchetto-Zolet AC, Pinheiro F, Salgueiro F, Palma-Silva C (2013) Phylogeographical patterns shed light on evolutionary process in South America. Mol Ecol 22:1193–1213. https://doi.org/10.1111/mec.12164
Turnbull WD (1970) Mammalian masticatory apparatus. Fieldiana Geology 18:149–356
Velasco JA, González-Salazar C (2019) Akaike information criterion should not be a “test” of geographical prediction accuracy in ecological niche modelling. Ecol Inform 51:25–32
Vieira EM, Astúa de Moraes D (2003) Carnivory and insectivory in Neotropical marsupials. In: Jones M, Dickman C, Archer M (eds). Predators With Pouches: The Biology of Carnivorous Marsupials. CSIRO Publishing, Victoria, pp 271–284
Voss RS, Emmons LH (1996) Mammalian diversity in Neotropical lowland rainforests: A preliminary assessment. Bull Amer Mus Nat Hist 230:1–115
Wible JR (2003) On the cranial osteology of the short-tailed opossum Monodelphis brevicaudata (Didelphidae, Marsupialia). Ann Carnegie Mus 72:137–202
Zachos J, Pagani M, Sloan L, Thomas E, Billups K (2001) Trends, rhythms, and aberrations in global climate 65 Ma to present. Science 292:686–693. https://doi.org/10.1126/science.105941
Zhang J, Kapli P, Pavlidis P, Stamatakis A (2013) A general species delimitation method with applications to phylogenetic placements. Bioinformatics 29:2869–2876. https://doi.org/10.1093/bioinformatics/btt499
Acknowledgements
We thank the curators for granting us access to the collections under their care: João Oliveira, Robert Voss, Pablo Teta, Raúl Maneyro, Sergio Bogan, Enrique González, Mario de Vivo, and Juliana Gualda. We thank Baltazar González for the discussion on ecological niche modelling (ENM), providing the code, and running the models. Facundo Morinigo and Alberto Fameli helped with the laboratory analyses. Adrian Monjeau and Nature Map Argentina provided the computing power needed for the ENM. Maria Claudene Barros and Daiane Chaves Nascimento sent us the sequences from their work in Brazil. GMM thanks Michael Simeon and Eugene Watkins for economic support. We also thank the two reviewers who helped to improve the manuscript.
Author information
Authors and Affiliations
Contributions
MAC conceived the study. MAC and RG-I performed the molecular analyses. GM performed the environmental niche modelling analyses. GM and MAC studied the morphology of the specimens. MAC and GM wrote the manuscript, MAC, RG-I and GM made the figures. RG-I reviewed and edited the text.All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Financial interests
The authors declare they have no financial interests. This work was supported by Agencia I + D + I from Argentina, grant numbers PICT 2016–3151 and PICT 2019 − 1545.
Competing interests
GMM is an Associate Editor of the Journal of Mammalian Evolution but was not involved in the evaluation of this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10914_2023_9685_MOESM1_ESM.xlsx
Online Resources 1-5, including sequence data (Online Resources 1-4) and list of specimens analyzed for the morphological analysis.
10914_2023_9685_MOESM3_ESM.pdf
Online Resources 7-13: Time-calibrated tree obtained from the analysis of the cyt b haplotypes (Online Resource 7),results of the phylogeographic analyses of haplogroups A and B (Online Resources 8-10), results of the speciesdelimitation analyses (Online Resources 11-12), and ecological niche models obtained for each group.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chemisquy, M.A., González-Ittig, R.E. & Martin, G.M. Hidden in plain sight: Didelphis albiventris (Didelphimorphia: Didelphidae) might not be a single species. J Mammal Evol 30, 873–889 (2023). https://doi.org/10.1007/s10914-023-09685-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10914-023-09685-1