Abstract
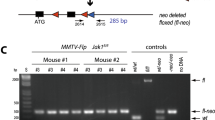
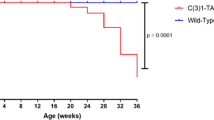
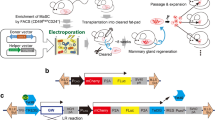
Genetically engineered mouse models have become an indispensable tool for breast cancer research. Combination of multiple site-specific recombination systems such as Cre/loxP and Flippase (Flp)/Frt allows for engineering of sophisticated, multi-layered conditional mouse models. Here, we report the generation and characterization of a novel transgenic mouse line expressing a mouse codon-optimized Flp under the control of the mouse mammary tumor virus (MMTV) promoter. These mice show robust Flp-mediated recombination in luminal mammary gland and breast cancer cells but no Flp activity in non-mammary tissues, with the exception of limited activity in salivary glands. These mice provide a unique tool for studying mammary gland biology and carcinogenesis in mice.



Similar content being viewed by others
Abbreviations
- MMTV:
-
Mouse mammary tumor virus
- Flp:
-
Flippase
- Flpo:
-
mouse codon usage-optimized Flp
- CK:
-
cytokeratin
References
Borowsky AD. Choosing a mouse model: experimental biology in context--the utility and limitations of mouse models of breast cancer. Cold Spring Harb Perspect Biol. 2011;3(9):a009670. https://doi.org/10.1101/cshperspect.a009670.
Van Keymeulen A, Lee MY, Ousset M, Brohee S, Rorive S, Giraddi RR, et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature. 2015;525(7567):119–23. https://doi.org/10.1038/nature14665.
Koren S, Reavie L, Couto JP, De Silva D, Stadler MB, Roloff T, et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature. 2015;525(7567):114–8. https://doi.org/10.1038/nature14669.
Blaas L, Pucci F, Messal HA, Andersson AB, Josue Ruiz E, Gerling M, et al. Lgr6 labels a rare population of mammary gland progenitor cells that are able to originate luminal mammary tumours. Nat Cell Biol. 2016;18(12):1346–56. https://doi.org/10.1038/ncb3434.
Zomer A, Ellenbroek SI, Ritsma L, Beerling E, Vrisekoop N, Van Rheenen J. Intravital imaging of cancer stem cell plasticity in mammary tumors. Stem Cells. 2013;31(3):602–6. https://doi.org/10.1002/stem.1296.
Beerling E, Seinstra D, de Wit E, Kester L, van der Velden D, Maynard C, et al. Plasticity between epithelial and mesenchymal states unlinks EMT from metastasis-enhancing stem cell capacity. Cell Rep. 2016;14(10):2281–8. https://doi.org/10.1016/j.celrep.2016.02.034.
Cheung KJ, Padmanaban V, Silvestri V, Schipper K, Cohen JD, Fairchild AN, et al. Polyclonal breast cancer metastases arise from collective dissemination of keratin 14-expressing tumor cell clusters. Proc Natl Acad Sci U S A. 2016;113(7):E854–63. https://doi.org/10.1073/pnas.1508541113.
Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527(7579):472–6. https://doi.org/10.1038/nature15748.
Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6(1):7–28.
Olorunniji FJ, Rosser SJ, Stark WM. Site-specific recombinases: molecular machines for the genetic revolution. Biochem J. 2016;473(6):673–84. https://doi.org/10.1042/BJ20151112.
Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. https://doi.org/10.1016/S0076-6879(10)77011-7.
Schonhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med. 2014;20(11):1340–7. https://doi.org/10.1038/nm.3646.
Shai A, Dankort D, Juan J, Green S, McMahon M. TP53 silencing bypasses growth arrest of BRAFV600E-induced lung tumor cells in a two-switch model of lung tumorigenesis. Cancer Res. 2015;75(15):3167–80. https://doi.org/10.1158/0008-5472.CAN-14-3701.
Young NP, Crowley D, Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71(11):4040–7. https://doi.org/10.1158/0008-5472.CAN-10-4563.
Moding EJ, Lee CL, Castle KD, Oh P, Mao L, Zha S, et al. Atm deletion with dual recombinase technology preferentially radiosensitizes tumor endothelium. J Clin Invest. 2014;124(8):3325–38. https://doi.org/10.1172/JCI73932.
He L, Li Y, Li Y, Pu W, Huang X, Tian X, et al. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat Med. 2017;23(12):1488–98. https://doi.org/10.1038/nm.4437.
Bang SJ, Jensen P, Dymecki SM, Commons KG. Projections and interconnections of genetically defined serotonin neurons in mice. Eur J Neurosci. 2012;35(1):85–96. https://doi.org/10.1111/j.1460-9568.2011.07936.x.
Engleka KA, Manderfield LJ, Brust RD, Li L, Cohen A, Dymecki SM, et al. Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ Res. 2012;110(7):922–6. https://doi.org/10.1161/CIRCRESAHA.112.266510.
Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12(3):954–61.
Lee CL, Moding EJ, Huang X, Li Y, Woodlief LZ, Rodrigues RC, et al. Generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. Dis Model Mech. 2012;5(3):397–402. https://doi.org/10.1242/dmm.009084.
Acknowledgments
We thank P. Lorentz for support with microscopy and H. Antoniadis and U. Schmieder for technical support. We are grateful to S. Dymecki and N. Hynes for providing mice and P. Soriano and K. Alitalo for plasmids. R.B. was supported by a MD-PhD fellowship of the Swiss National Science Foundation and Swiss Cancer League. The work was supported by the Swiss National Science Foundation and the Swiss Cancer League.
Author information
Authors and Affiliations
Contributions
F.L. designed and performed the experiments, analysed the data and wrote the manuscript. R.B. conceived the idea and designed some of the experiments and analysed the data. A.V. performed some of the experiments. H.O. and P.P. performed the pronuclei microinjections. G.C. supervised the project, designed the experiments, analysed the data and wrote the manuscript.
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare no financial and non-financial competing interests.
Ethical Approval
All animal experiments were carried out in accordance with the guidelines of the Swiss Federal Veterinary Office (SFVO) and the Cantonal Veterinary Office of Basel-Stadt (license numbers 1878, 1023G1).
Informed Consent
No individual person’s data has been used in this project.
Rights and permissions
About this article
Cite this article
Lüönd, F., Bill, R., Vettiger, A. et al. A Transgenic MMTV-Flippase Mouse Line for Molecular Engineering in Mammary Gland and Breast Cancer Mouse Models. J Mammary Gland Biol Neoplasia 24, 39–45 (2019). https://doi.org/10.1007/s10911-018-9412-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-018-9412-4