Abstract
The noncanonical NF-κB pathway is implicated in diverse biological and immunological processes. Monoallelic C-terminus loss-of-function and gain-of-function mutations of NFKB2 have been recently identified as a cause of immunodeficiency manifesting with common variable immunodeficiency (CVID) or combined immunodeficiency (CID) phenotypes. Herein we report a family carrying a heterozygous nonsense mutation in NFKB2 (c.809G > A, p.W270*). This variant is associated with increased mRNA decay and no mutant NFKB2 protein expression, leading to NFKB2 haploinsufficiency. Our findings demonstrate that bona fide NFKB2 haploinsufficiency, likely caused by mutant mRNA decay and protein instability leading to the transcription and expression of only the wild-type allele, is associated with clinical immunodeficiency, although with incomplete clinical penetrance. Abnormal B cell development, hypogammaglobulinemia, poor antibody response, and abnormal noncanonical (but normal canonical) NF-κB pathway signaling are the immunologic hallmarks of this disease. This adds a third allelic variant to the pathophysiology of NFKB2-mediated immunodeficiency disorders.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) is a major transcription factor which plays an important role in the immune system. The NF-κB family has five members, RelA, RelB, c-Rel, NFΚB1-p105/p50, and NFΚB2-p100/p52, and they share an N-terminal Rel homology domain (RHD) required for DNA binding and homo/hetero dimerization [1]. NFKB2 is translated as a precursor protein p100, contains an IκB-like-C-terminal portion, and inhibits the nuclear translocation of associated NF-κB members. Proteasome-mediated processing of p100, which is tightly regulated in a signal-induced manner, produces active form of p52 which leads to nuclear translocation of NF-κB complex, predominantly RelB/p52, and regulates target gene expression [2, 3].
Human NFKB2 gene defects have been shown to be associated with B cell dysregulation in patients with common variable immunodeficiency (CVID) or combined immunodeficiency (CID) [4]. CVID with endocrinopathy and ectodermal dysplasia and CID phenotypes have been associated with NFKB2 C-terminus loss-of-function (LOF) mutations, which disrupt the phosphorylation and block the processing of p100 to p52, resulting in cytoplasmic retention of mutant p100 and reduction of p52 expression and nuclear translocation [4, 5]. More recently, our group reported NFKB2 gain-of-function (GOF) mutations in CID patients with B cell lymphopenia, hypogammaglobulinemia, and multiple severe infections (i.e., Pneumocystis jirovecii pneumonitis, EBV, and CVM viremia, among others) with incomplete penetrance [6]. In vitro studies using mutant constructs showed that GOF mutants (E418* and R635*) translocated to the nucleus with RelB in a NF-κB-inducing kinase (NIK)-independent manner, increased CXCL13 gene transcription, and also increased canonical NF-κB activity [6]. Herein, we describe three members of a family carrying a heterozygous nonsense NFKB2 mutation that causes p100 and p52 haploinsufficiency through mRNA decay and/or protein instability, presenting as predominantly antibody deficiency in one individual (II.2).
Materials and Methods
Human Subjects
All patients or their guardians provided written informed consent in accordance with the Declaration of Helsinki under institutional review board−approved protocols of the National Institute of Allergy and Infectious Diseases, NIH. Blood from healthy donors and patients was obtained under approved protocols, which also allowed for the collection and use of patients’ family history and pedigrees for publication. All procedures were based on standard of care, and established clinical guidelines were followed.
Next-Generation Sequencing
For the proband (II.2), whole exome sequencing (WES) was performed using the Ion Torrent AmpliSeq RDY Exome Kit and the Ion Chef and Proton instruments (Life Technologies). For family members (I.1 and II.1), WES was performed using the Nextera DNA Exome Kit and the HiSeq 2500 instrument (Illumina). All WES experiments were performed according to the manufacturer’s protocols.
Sanger Sequencing
Sanger sequencing was performed to confirm WES-detected variants and to screen family members as described previously [6]. For NFKB2 sequencing in cDNA, total RNA was isolated from PBMCs using the RNeasy Plus mini kit (Qiagen) and reverse transcribed by the use of the QuantiTech reverse transcription kit (Qiagen). cDNA was PCR-amplified using GoTaq polymerase (Promega) and exon-specific NFKB2 primers. Forwards: 5’-CTAGACAGAGCCGGGCCTAG-3′ or 5’-CTCTAGGCCCCAGGGCCTTA-3′.
; reverse: 5’-CAGGTGGCGCTGTCCCGCCA-3′.
Immunoblotting
Total PBMCs were stimulated with anti-CD3 (OKT3, 1μg/mL; eBioscience; 16–0037-85) for 48 h. Cells were washed with PBS once and lysed in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100 and 0.5% NP40 and halt protease and phosphatase inhibitor cocktail [Sigma]), and samples were adjusted to have equal concentration of total protein and subjected to immunoblotting. Cell lysates were prepared and analyzed with anti-phospho-NFKB2 (S866/S870, Cell signaling #4810) and anti-NFKB2 (Cell Signaling Technology #3017) as previously described [6]. For the T cells blasts, total PBMCs were stimulated with anti-CD3 and anti-CD28 (1μg/mL; eBioscience) in the presence of IL-2 (10 ng/ml, Cell Signaling Technology) and cultured for 6–10 days. IL-2 was added every 2–3 days.
Evaluation of Cell Proliferation
PBMCs, either freshly isolated or thawed from liquid nitrogen-stored samples, were incubated with CellTrace Violet Cell Proliferation Kit (1 μM; Invitrogen). After 20 min, 10 volumes of RPMI/10% fetal bovine serum (FBS) were added, and the cells were washed twice with RPMI/10% FBS. 1 × 105 cells were seeded into 96-well plates and stimulated with anti-CD3 antibody and anti-CD28 antibody (1 μg/mL each, eBioscience; 16–0037-85 and 16–0289-85). For B cell proliferation, cells were stimulated with anti-IgM (10 μg/ml, Jackson Lab; 109–006-129), CD40L (100 ng/ml, Enzo; ALX522–110-C010), CpG (0.5 μM, Enzo; ALX 746–006-C100), IL-4 (50 ng/ml, Peprotech; 200–04), and IL-21 (50 ng/ml, Peprotech; 200–21) as indicated in the figure. After incubation for 3–5 days (3 days for T cells and 4–5 days for B cells), cells were stained with fluorochrome-conjugated CD4, CD8, or CD19 (BD Biosciences, Clone RPA-T4, RPA-T8, HIB19 respectively) for 30 min at 4 °C (dark). Cells were washed with PBS twice, and cells were acquired and analyzed by flow cytometry (Becton Dickinson FACSCanto II) and FlowJo software (FlowJo 10.5.2, TreeStar).
Results
Index patient II.2 is a 14-year-old Hispanic boy diagnosed with CVID at the age of 7 years. He was born at term to a young, non-consanguineous family. At the age of 1 month, he presented with bronchiolitis, his first infectious episode. He was diagnosed with a left lobar pneumonia with effusion at the age of 2 years and a right lobar pneumonia at age 6 (no organisms identified). He received all scheduled vaccines without complications. Clinical and laboratory immune evaluation at age 7 was relevant for clubbing, difficulty breathing, and a chest CT scan showing bilateral bronchiectasis and segmental atelectasis. His immunologic laboratory evaluation showed hypogammaglobulinemia involving the 3 major isotypes with IgG 472 mg/dL, IgA < 7 mg/dL, and IgM 21 mg/dL; protective titers to tetanus toxoid, but negative to measles, mumps, rubella, hepatitis A, and hepatitis B; low CD3 and CD4 T cells, with normal CD8 T cell, NK cell, and B cell enumeration; the patient also presented abnormal B cell subsets with low memory B cells (patient 8) [7] and reduced somatic hypermutation (patient 14) [8] as previously reported. Since his diagnosis the patient has been receiving IgG replacement and antibiotic prophylaxis with partial reduction in his respiratory infection rate. Immune evaluation at age 14 showed similar laboratory results (Table 1). His father (I.1) and sister (II.1) are healthy and clinically asymptomatic.
Whole exome sequence (WES) and/or Sanger confirmation was performed in all family members. A heterozygous nonsense NFKB2 mutation creating a premature stop codon p.W270* (NM_001077494, c.809G > A, p.W270*) was detected in the proband II.2, his sister II.1, and their father I.1 (Fig. 1a–c, Table 2). This variant was predicted to undergo nonsense-mediated decay by MutationTaster and to be deleterious by the Combined Annotation Dependent Depletion algorithm (CADD GRCh37-v1.4), with CADD phred scores of 37, where CADD_phred scores > 20 predict that a variant is among the 1% most deleterious of all possible substitutions. Variant c.809G > A was not reported in the Genome Aggregation Database (gnomAD; gnomad.broadinstitute.org), a database that generally excludes individuals with severe pediatric diseases.The sequencing data have been deposited with links to BioProject accession number PRJNA655454 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
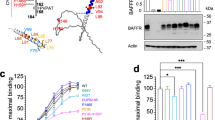
Genetic and pedigree of family with NFKB2 mutation. (a) Pedigree showing family members with a germline mutation in NFKB2. Affected or asymptomatic mutation positive individuals indicated by filled black color or gray color respectively. (b) Schematic representation of the domains of NFKB2-p100 showing Rel homology domain (RHD), ankyrin repeats (ARD), death domain (DD), and location of mutations. Previously reported germline NFKB2 mutations in CVID or CID patients are indicated below the domain. Blue color indicates gain-of-function (GOF; E418* and R635*); C-terminus loss-of-function mutations are shown in red (LOF; R853fs, R853X, D854fs, K855fs, K855X, D865fs, D865G, S866R, S866fs, A867fs, A867V, Y868C, and Q871X). (c) Sequence chromatogram of genomic DNA (from whole blood) or complementary DNA (from total PBMCs) from indicated individuals with NFKB2 mutation. Arrows indicate the position of the mutation
While NF-κB canonical activation is rapid and independent of protein synthesis, the noncanonical NF-κB pathway is slow and relies on the inducible processing of NFKB2 precursor. Previous reports demonstrated that TCR signaling can stimulate the NF-κB noncanonical pathway, as other well-known agonists (e.g., CD40L, lymphotoxin, RANKL, and BAFF) also do [9]. To induce the activation of the noncanonical NF-κB pathway, PBMCs of all mutation-positive family members, as well as healthy donors (HD), were stimulated with anti-CD3 for 48 h, and NFΚΒ2 protein expression was tested. Immunoblot analysis revealed that PBMCs expressed markedly reduced levels of p100 and p52 as well as phosphorylated NFKΒ2/p100 upon stimulation, when compared to healthy donor controls. Truncated mutant NFKB2 p.W270* (predicted at 30 kDa) was not detected in the three mutation carriers (Fig. 2a). Due to the different lymphocyte numbers and subsets between the three mutation carriers, T cells blasts were generated to test NFKB2 expression in TCR-expressing/responsive cells. The mutant protein was not detected either in the T cell blasts, similar as in the PBMC experiment. Interestingly, the clinically affected family member (II.2) showed a markedly reduced NFKB2 p100/p52 expression when compared with the asymptomatic mutation carriers or the healthy donor controls (Fig. 2b). Based on the above-mentioned results, we speculated the mutant protein could be unstable, its mRNA subject to decay, or both mechanisms be involved. NFKB2 mRNA to cDNA conversion and further sequencing revealed that while the affected patient (II.2) and his father’s (I.1) cells only transcribed the wild type allele; the sister’s (II.1) cells transcribed low/residual amounts of the mutant allele when compared with the wild-type allele (Fig. 1c). To further test the effect of reduced NFΚΒ2 expression on downstream signaling, CXCL13 expression, a chemokine associated with noncanonical NF-κB activation, was measured by real-time PCR assay. Both, patient II.2 and his asymptomatic father I.1 showed markedly reduced CXCL13 expression, suggesting that NFΚΒ2 haploinsufficiency also affects downstream signaling, although it does not correlate with clinical disease penetrance (Fig. 3; sister II.1 was not tested due to limited sample availability). The canonical NF-κB signaling, which showed increased cytokine generation upon lipopolysaccharide (LPS) stimulation in patients carrying NFKB2 GOF mutations [6], was comparable with the HD in I.1 and II.2 (Fig. 3b–c).
NFKB2 protein expression in PBMCs and T cell blasts. (a) Total PBMCs from healthy donors (HD) and individuals with NFKB2 mutation were stimulated with anti-CD3 (OKT3, 1ug/ml) for 48h. Cell lysates were prepared and analyzed for the phosphorylation of p100 (S866/870, Cell signaling #4810) and expression of full-length (p100) and processed form (p52) by western blotting (Cell signaling #3017) as previously described [6]. b-actin was used as a loading control. Data shown are representative of two independent experiments. (b) To generate T cell blasts, total PBMCs were stimulated with anti-CD3 and anti-CD28 (1 ug/mL) in the presence of IL-2 (10ng/ml) for 6-10 days. IL-2 was added every 2-3 days. Cell lysates were prepared and analyzed for the full-length (p100) and processed form (p52) of NF-kB by western blotting. b-actin was used as a loading control. Data shown are representative of two independent experiments. The graphs show the relative expression levels of p100 and p52, determined with densitometry and normalized to b-actin protein expression using Image Studio
CXCL13 expression and cytokine generation in patients with NFKB2 mutation. (a) Enriched CD4 T cells were stimulated with Dynabeads-CD3/28 for 48 h. Levels of CXCL13 mRNA in activated CD4 T cells were measured by real-time PCR using the probe for CXCL13 and normalized to 18S rRNA as previously described [6]. Data obtained from two independent experiments with two normal controls in each experiment. All data were performed in triplicate and the expression of CXCL13 in patient samples was relatively quantified by comparing with the average value of each set of normal controls, with expression levels normalized as 1. Data are shown as means +SEM of four healthy donor controls (HD), I.1 (n = 1) and II.2 (n = 2). ‘n’ represents the number of replicates. (b-c) Total PBMCs were stimulated with LPS (100 ng/ml) for 24 h. Cytokine expression in supernatants was analyzed by using multiplex bead assay (R&D systems) and Luminex 100/200 according to the manufacture’s instruction. Data are shown as means +SEM of three healthy donor controls (HD)
When T and B cell proliferation were tested in the index patient and his father, normal TCR-induced T cell proliferation and normal B cell proliferation upon T cell-dependent and independent activation were observed, despite the changes observed in their immune phenotype (Table 1) (Fig. 4a–b).
Proliferation of T and B cells from patients and healthy donor controls. (a) Total PBMCs were stained with CellTrace Violet and stimulated with anti-CD3 and anti-CD28 for 3 days. Data are representative of three experiments. (b) For B cell proliferation, cells were stimulated with indicated agonists (anti-IgM, CD40L, IL-4, CpG, IL-21) for 4–5 days. Numbers indicate the percentage of cells having undergone at least one cellular division assessed by CellTrace Violet dye dilution. Data are representative of two experiments
Altogether these results suggest that (a) the mutant mRNA is fully (i.e., I.1 and II.2) or partially (i.e., II.1) unstable and degraded through mechanisms like nonsense-mediated mRNA-decay (NMD) (Fig. 1a) and (b) the transcribed mutant protein, if any, is unstable and undetectable (Fig. 2). Moreover, low NFΚΒ2 expression in the symptomatic patient’s T cell blasts (~50% reduction in p100 and > 90% reduction in p52 compared with the normal controls) compared with the asymptomatic mutation carriers (15–20% reduction in p100 and 30–60% in p52) may help to explain the clinical penetrance correlation, but further research is needed to support this finding.
Discussion
In this study we demonstrate NFKB2 haploinsufficiency in a patient with a pediatric onset CVID phenotype. The product of the mutated allele is deficient either because of mRNA decay and/or protein degradation, resulting in reduction of both p100 and p52 NFKB2 protein expression and NFKB2/p100 phosphorylation upon stimulation. This is a different mechanism than NFKB2 C-terminus LOF mutations originally reported by Chen et al., (so-called p52 haploinsufficiency) [5] or GOF mutations originally reported by our group [6]. While NFKB2 C-terminus LOF mutations are associated with reduced p52 expression and nuclear translocation due to the failure of phosphorylation and processing of C-terminus-mutated p100; GOF mutations show decreased levels of p100 due to truncated mutant expression leading to increased nuclear translocation of RelB/GOF mutant complexes that in its turn increase the activation of downstream pathways [6]. Moreover, while C-terminal LOF mutations are almost fully penetrant (1/43 asymptomatic) and endocrine defects are common (21/43), GOF and haploinsufficiency mutations are not fully penetrant (1 (or 2)/5 asymptomatic for GOF and 2/3 for haploinsufficiency) and endocrine defects have not been described in neither of these allelic variants [4,5,6]. It is likely that the C-terminus NFKB2 mutants exert a dominant negative effect on NF-kB signaling by preventing nuclear translocation of associated NF-κB members, that in its turn results in an almost fully penetrant disease.
Of note, somatic NFKB2 gene arrangements producing partially deleted C-terminal domains which lost transcriptional repression activity and facilitated its constitutive transcriptional activity have been shown to be associated with human malignant hematologic diseases [10]. Four cases of malignancies were also reported in patients with germline NFKB2 mutations, one patient with GOF mutation and three in patients with C-terminus LOF mutations, providing evidence that aberrant or constitutive NFKB2 activation disrupts the normal cell function and could lead to cancer development as well as immune dysregulation [4].
CVID is the most common symptomatic phenotype among primary immunodeficiency patients, and monogenic cases account for ~10–20% of them [11]. Several reports demonstrated that the noncanonical NF-κB pathway has a critical role in B cell differentiation and isotype class switching [3, 12] and several variants in noncanonical pathway have been reported in CVID patients such as BAFFR, TACI, TRAF3, NIK, and NFKB2 [4, 11]. Of note, in 2012 patient II.2 was reported to have 2 synonymous TACI variants, homozygous p.T27T (rs8072293) and heterozygous p.S277S (rs11078355) [7]. These variants are now considered to be likely benign and present in high proportions in different healthy control populations (allele frequency 0.81 and 0.49, respectively; gnomAD). Moreover, the asymptomatic father I.1 and sister II.1 in this report carry the same TACI synonymous variants in the same zygosity status as the index patient II.2 and therefore are unlikely associated with disease clinical penetrance or expressivity by themselves. Most of previously reported NFKB2-mutated patients have abnormal B cell differentiation with reduced switched memory B cells and some with low NK cell number [4]. Similar as previous findings, low-switched memory B cells and hypogammaglobulinemia were observed in patient II.2 (Table 1), suggesting that B cell development was dysregulated in the patient with NFKB2 haploinsufficiency.
Interestingly, a female patient with a novel heterozygous interstitial 5.54 Mb deletion at 10q24.31-q25.1 was previously reported [13]. This defect involved 90+ genes, including NFKB2, and the patient presented at birth with lobar holoprosencephaly, cleft lip and palate, and hypoplastic kidneys, among multiple congenital anomalies. As the clinical and laboratory immunologic phenotype was not part of the original description, and the patient could not be evaluated for our study, the role of NFKB2 haploinsufficiency in her phenotype remains to be determined. Noteworthy, patients with chromosomal aberrations could provide important clues for the study of primary immunodeficiencies [14].
In summary, NFΚΒ2 haploinsufficiency appears as a novel pathophysiologic mechanism underlying primarily antibody deficiency as CVID, with incomplete clinical penetrance. Our findings extend the genetic, immunologic, and pathophysiology underlying NFKB2-mediated disease, adding a new allelic variant (haploinsufficiency) to the previously reported mechanisms (i.e., C-terminus LOF, and GOF) and contributing to the penetrance-expressivity conundrum associated with these defects.
Change history
09 September 2020
Due to typesetting mistake, the caption of Figure 2 was mistakenly replaced with the caption of Figure 3.
References
Zhang Q, Lenardo MJ, Baltimore D. 30 years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell. 2017;168:37–57.
Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell Res. 2011;21:223–44.
Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85.
Klemann C, Camacho-Ordonez N, Yang L, Eskandarian Z, Rojas-Restrepo JL, Frede N, et al. Clinical and immunological phenotype of patients with primary immunodeficiency due to damaging mutations in NFKB2. Front Immunol. 2019;10:297.
Chen K, Coonrod EM, Kumanovics A, Franks ZF, Durtschi JD, Margraf RL, et al. Germline mutations in NFKB2 implicate the noncanonical NF-kappaB pathway in the pathogenesis of common variable immunodeficiency. Am J Hum Genet. 2013;93:812–24.
Kuehn HS, Niemela JE, Sreedhara K, Stoddard JL, Grossman J, Wysocki CA, et al. Novel nonsense gain-of-function NFKB2 mutations associated with a combined immunodeficiency phenotype. Blood. 2017;130:1553–64.
Almejun MB, Sajaroff E, Galicchio M, Oleastro M, Bernasconi A, Zelazko M, et al. Immunological characteristics and two novel mutations in TACI in a cohort of 28 pediatric patients with common variable immunodeficiency. J Clin Immunol. 2012;32:89–97.
Almejun MB, Campos BC, Patino V, Galicchio M, Zelazko M, Oleastro M, et al. Noninfectious complications in patients with pediatric-onset common variable immunodeficiency correlated with defects in somatic hypermutation but not in class-switch recombination. J Allergy Clin Immunol. 2017;139:913–22.
Michel M, Wilhelmi I, Schultz AS, Preussner M, Heyd F. Activation-induced tumor necrosis factor receptor-associated factor 3 (Traf3) alternative splicing controls the noncanonical nuclear factor kappaB pathway and chemokine expression in human T cells. J Biol Chem. 2014;289:13651–60.
Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–47.
Bogaert DJ, Dullaers M, Lambrecht BN, Vermaelen KY, De Baere E, Haerynck F. Genes associated with common variable immunodeficiency: one diagnosis to rule them all? J Med Genet. 2016;53:575–90.
Rickert RC, Jellusova J, Miletic AV. Signaling by the tumor necrosis factor receptor superfamily in B-cell biology and disease. Immunol Rev. 2011;244:115–33.
Peltekova IT, Hurteau-Millar J, Armour CM. Novel interstitial deletion of 10q24.3–25.1 associated with multiple congenital anomalies including lobar holoprosencephaly, cleft lip and palate, and hypoplastic kidneys. Am J Med Genet A. 2014;164A:3132–6.
Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, et al. Loss of B Cells in Patients with Heterozygous Mutations in IKAROS. N Engl J Med. 2016;374:1032–43.
Garcia-Prat M, Alvarez-Sierra D, Aguilo-Cucurull A, Salgado-Perandres S, Briongos-Sebastian S, Franco-Jarava C, et al. Extended immunophenotyping reference values in a healthy pediatric population. Cytometry B Clin Cytom. 2019;96:223–33.
Acknowledgments
We thank the family evaluated in this work for their contributions to the study. This research was supported by the Intramural Research Program, NIH Clinical Center, US National Institutes of Health (NIH). and a grant from the Colombian Administrative Department of Science, Technology and Innovation Colciencias (111556934592, contract 569-2013). The content of this article does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors have no conflict of interest to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original version of this article was revised: The caption of Figure 2 was incorrect.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Kuehn, H.S., Bernasconi, A., Niemela, J.E. et al. A Nonsense N –Terminus NFKB2 Mutation Leading to Haploinsufficiency in a Patient with a Predominantly Antibody Deficiency. J Clin Immunol 40, 1093–1101 (2020). https://doi.org/10.1007/s10875-020-00842-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-020-00842-2