Abstract
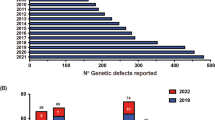
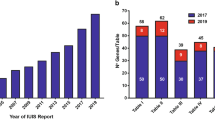
Primary immunodeficiency diseases (PIDs) comprise a group of highly heterogeneous immune system diseases and around 300 forms of PID have been described to date. Next Generation Sequencing (NGS) has recently become an increasingly used approach for gene identification and molecular diagnosis of human diseases. Herein we summarize the practical considerations for the interpretation of NGS data and the techniques for searching disease-related PID genes, and suggest future directions for research in this field.


Similar content being viewed by others
Abbreviations
- NGS:
-
Next generation sequencing
- PID:
-
Primary immunodeficiency diseases
- MDs:
-
Mendelian disorders
- WGS:
-
Whole genome sequencing
- WES:
-
Whole exome sequencing
- QC:
-
Quality control
References
Routes J, Abinun M, Al-Herz W, Bustamante J, Condino-Neto A, De La Morena MT, et al. ICON: the early diagnosis of congenital immunodeficiencies. Journal of Clinical Immunology. 2014;34(4):398–424.
Joshi AY, Iyer VN, Hagan JB, St Sauver JL, Boyce TG. Incidence and temporal trends of primary immunodeficiency: a population-based cohort study. Mayo Clinic Proceedings. 2009;84(1):16–22.
Hammarstrom L, Vorechovsky I, Webster D. Selective IgA deficiency (SIgAD) and common variable immunodeficiency (CVID). Clinical and Experimental Immunology. 2000;120(2):225–31.
van den Berg JM, van Koppen E, Ahlin A, Belohradsky BH, Bernatowska E, Corbeel L, et al. Chronic granulomatous disease: the European experience. PLoS ONE. 2009;4(4):e5234.
Bousfiha AA, Jeddane L, Ailal F, Benhsaien I, Mahlaoui N, Casanova JL, et al. Primary immunodeficiency diseases worldwide: more common than generally thought. Journal of Clinical Immunology. 2013;33(1):1–7.
Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, et al. Primary Immunodeficiency Diseases: an Update on the Classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. Journal of Clinical Immunology. 2015.
Bousfiha A, Jeddane L, Al-Herz W, Ailal F, Casanova JL, Chatila T, et al. The 2015 IUIS phenotypic classification for primary immunodeficiencies. Journal of Clinical Immunology. 2015;35(8):727–38.
Itan Y, Casanova JL. Novel primary immunodeficiency candidate genes predicted by the human gene connectome. Frontiers in Immunology. 2015;6:142.
Germeshausen M, Zeidler C, Stuhrmann M, Lanciotti M, Ballmaier M, Welte K. Digenic mutations in severe congenital neutropenia. Haematologica. 2010;95(7):1207–10.
Zhang K, Chandrakasan S, Chapman H, Valencia CA, Husami A, Kissell D, et al. Synergistic defects of different molecules in the cytotoxic pathway lead to clinical familial hemophagocytic lymphohistiocytosis. Blood. 2014;124(8):1331–4.
Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–6.
Roach JC, Glusman G, Smit AF, Huff CD, Hubley R, Shannon PT, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science (New York, NY). 2010;328(5978):636–9.
Wang JL, Yang X, Xia K, Hu ZM, Weng L, Jin X, et al. TGM6 identified as a novel causative gene of spinocerebellar ataxias using exome sequencing. Brain: a Journal of Neurology. 2010;133(Pt 12):3510–8.
Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, et al. Exome sequencing identifies the cause of a mendelian disorder. Nature Genetics. 2010;42(1):30–5.
Lupski JR, Reid JG, Gonzaga-Jauregui C, Rio Deiros D, Chen DC, Nazareth L, et al. Whole-genome sequencing in a patient with Charcot-Marie-Tooth neuropathy. The New England Journal of Medicine. 2010;362(13):1181–91.
Bonnefond A, Durand E, Sand O, De Graeve F, Gallina S, Busiah K, et al. Molecular diagnosis of neonatal diabetes mellitus using next-generation sequencing of the whole exome. PLoS ONE. 2010;5(10):e13630.
Notarangelo LD. Primary immunodeficiencies. The Journal of Allergy and Clinical Immunology. 2010;125(2 Suppl 2):S182–94.
Alkhairy OK, Perez-Becker R, Driessen GJ, Abolhassani H, van Montfrans J, Borte S, et al. Novel mutations in TNFRSF7/CD27: clinical, immunologic, and genetic characterization of human CD27 deficiency. The Journal of Allergy and Clinical Immunology. 2015.
Abolhassani H, Cheraghi T, Rezaei N, Aghamohammadi A, Hammarstrom L. Common variable immunodeficiency or late-onset combined immunodeficiency: A new hypomorphic JAK3 patient and review of the literature. Journal of Investigational Allergology & Clinical Immunology. 2015;25(3):218–20.
Alkhairy OK, Abolhassani H, Rezaei N, Fang M, Andersen KK, Chavoshzadeh Z, et al. Spectrum of Phenotypes Associated with Mutations in LRBA. Journal of clinical immunology. 2015.
Abolhassani H, Wang N, Aghamohammadi A, Rezaei N, Lee YN, Frugoni F, et al. A hypomorphic recombination-activating gene 1 (RAG1) mutation resulting in a phenotype resembling common variable immunodeficiency. The Journal of Allergy and Clinical Immunology. 2014;134(6):1375–80.
Alkhairy OK, Rezaei N, Graham RR, Abolhassani H, Borte S, Hultenby K, et al. RAC2 loss-of-function mutation in 2 siblings with characteristics of common variable immunodeficiency. The Journal of allergy and clinical immunology. 2015;135(5):1380–4.e1–5.
Volk T, Pannicke U, Reisli I, Bulashevska A, Ritter J, Bjorkman A, et al. DCLRE1C (ARTEMIS) mutations causing phenotypes ranging from atypical severe combined immunodeficiency to mere antibody deficiency. Human Molecular Genetics. 2015;24(25):7361–72.
Salzer U, Bacchelli C, Buckridge S, Pan-Hammarstrom Q, Jennings S, Lougaris V, et al. Relevance of biallelic versus monoallelic TNFRSF13B mutations in distinguishing disease-causing from risk-increasing TNFRSF13B variants in antibody deficiency syndromes. Blood. 2009;113(9):1967–76.
Ferreira RC, Pan-Hammarstrom Q, Graham RR, Gateva V, Fontan G, Lee AT, et al. Association of IFIH1 and other autoimmunity risk alleles with selective IgA deficiency. Nature Genetics. 2010;42(9):777–80.
McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics (Oxford, England). 2010;26(16):2069–70.
Guo Y, Zhao S, Lehmann BD, Sheng Q, Shaver TM, Stricker TP, et al. Detection of internal exon deletion with exon del. BMC Bioinformatics. 2014;15:332.
Shi Y, Majewski J. FishingCNV: a graphical software package for detecting rare copy number variations in exome-sequencing data. Bioinformatics. 2013;29(11):1461–2.
Keerthikumar S, Raju R, Kandasamy K, Hijikata A, Ramabadran S, Balakrishnan L, et al. RAPID: resource of Asian primary immunodeficiency diseases. Nucleic Acids Research. 2009;37(Database issue):D863–7.
Casanova JL, Conley ME, Seligman SJ, Abel L, Notarangelo LD. Guidelines for genetic studies in single patients: lessons from primary immunodeficiencies. The Journal of Experimental Medicine. 2014;211(11):2137–49.
Lamperti C, Fang M, Invernizzi F, Liu X, Wang H, Zhang Q, et al. A novel homozygous mutation in SUCLA2 gene identified by exome sequencing. Molecular Genetics and Metabolism. 2012;107(3):403–8.
Hu H, Roach JC, Coon H, Guthery SL, Voelkerding KV, Margraf RL, et al. A unified test of linkage analysis and rare-variant association for analysis of pedigree sequence data. Nature Biotechnology. 2014;32(7):663–9.
Koboldt DC, Larson DE, Wilson RK. Using VarScan 2 for Germline Variant Calling and Somatic Mutation Detection. Current protocols in bioinformatics/editoral board, Andreas D Baxevanis [et al]. 2013;44:15.4.1–.4.7.
Moreau Y, Tranchevent LC. Computational tools for prioritizing candidate genes: boosting disease gene discovery. Nature Reviews Genetics. 2012;13(8):523–36.
Yang H. Robinson PN. Phenolyzer: Phenotype-Based Prioritization of Candidate Genes for Human Diseases. 2015;12(9):841–3.
Javed A, Agrawal S, Ng PC. Phen-gen: combining phenotype and genotype to analyze rare disorders. Nature Methods. 2014;11(9):935–7.
Shyr C, Tarailo-Graovac M, Gottlieb M, Lee JJ, van Karnebeek C, Wasserman WW. FLAGS, frequently mutated genes in public exomes. BMC Medical Genomics. 2014;7:64.
Itan Y, Shang L, Boisson B, Patin E, Bolze A, Moncada-Velez M, et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proceedings of the National Academy Of Sciences of the United States Of America. 2015;112(44):13615–20.
Vorechovsky I, Luo L, Hertz JM, Froland SS, Klemola T, Fiorini M, et al. Mutation pattern in the Bruton’s tyrosine kinase gene in 26 unrelated patients with X-linked agammaglobulinemia. Human Mutation. 1997;9(5):418–25.
Conley ME, Mathias D, Treadaway J, Minegishi Y, Rohrer J. Mutations in btk in patients with presumed X-linked agammaglobulinemia. American Journal of Human Genetics. 1998;62(5):1034–43.
Holinski-Feder E, Weiss M, Brandau O, Jedele KB, Nore B, Backesjo CM, et al. Mutation screening of the BTK gene in 56 families with X-linked agammaglobulinemia (XLA): 47 unique mutations without correlation to clinical course. Pediatrics. 1998;101(2):276–84.
Valiaho J, Faisal I, Ortutay C, Smith CI, Vihinen M. Characterization of all possible single-nucleotide change caused amino acid substitutions in the kinase domain of Bruton tyrosine kinase. Human Mutation. 2015;36(6):638–47.
Zeng H, Tao Y, Chen X, Zeng P, Wang B, Wei R, et al. Primary immunodeficiency in South China: clinical features and a genetic subanalysis of 138 children. Journal of Investigational Allergology & Clinical Immunology. 2013;23(5):302–8.
Lee WI, Torgerson TR, Schumacher MJ, Yel L, Zhu Q, Ochs HD. Molecular analysis of a large cohort of patients with the hyper immunoglobulin M (IgM) syndrome. Blood. 2005;105(5):1881–90.
Clark MJ, Chen R, Lam HY, Karczewski KJ, Chen R, Euskirchen G, et al. Performance comparison of exome DNA sequencing technologies. Nature Biotechnology. 2011;29(10):908–14.
Acknowledgments
We appreciate the participation of the patients and their families; without their support, this work would not have been possible.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Authorship Contributions
M.F. performed bioinformatics analysis, results interpretation, made the figures and wrote the paper; H.A. performed experiments, analyzed results and wrote the paper; C.K. analyzed results and wrote the paper. J. Z. performed the WES and L.H. designed the project and edited the paper.
Disclosure of Conflicts of Interest
None.
Rights and permissions
About this article
Cite this article
Fang, M., Abolhassani, H., Lim, C.K. et al. Next Generation Sequencing Data Analysis in Primary Immunodeficiency Disorders – Future Directions. J Clin Immunol 36 (Suppl 1), 68–75 (2016). https://doi.org/10.1007/s10875-016-0260-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10875-016-0260-y