Abstract
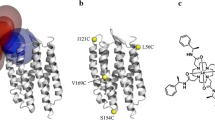
A single experimental method alone often fails to provide the resolution, accuracy, and coverage needed to model integral membrane proteins (IMPs). Integrating computation with experimental data is a powerful approach to supplement missing structural information with atomic detail. We combine RosettaNMR with experimentally-derived paramagnetic NMR restraints to guide membrane protein structure prediction. We demonstrate this approach using the disulfide bond formation protein B (DsbB), an α-helical IMP. Here, we attached a cyclen-based paramagnetic lanthanide tag to an engineered non-canonical amino acid (ncAA) using a copper-catalyzed azide-alkyne cycloaddition (CuAAC) click chemistry reaction. Using this tagging strategy, we collected 203 backbone HN pseudocontact shifts (PCSs) for three different labeling sites and used these as input to guide de novo membrane protein structure prediction protocols in Rosetta. We find that this sparse PCS dataset combined with 44 long-range NOEs as restraints in our calculations improves structure prediction of DsbB by enhancements in model accuracy, sampling, and scoring. The inclusion of this PCS dataset improved the Cα-RMSD transmembrane segment values of the best-scoring and best-RMSD models from 9.57 Å and 3.06 Å (no NMR data) to 5.73 Å and 2.18 Å, respectively.





Similar content being viewed by others
References
Chen JL et al (2016) Analysis of the solution conformations of T4 lysozyme by paramagnetic NMR spectroscopy. Phys Chem Chem Phys 18:5850–5859
Chin JW et al (2002) Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. J Am Chem Soc 124:9026–9027
Crick DJ et al (2015) Integral membrane protein structure determination using pseudocontact shifts. J Biomol NMR 61:197–207
de la Cruz L et al (2011) Binding of low molecular weight inhibitors promotes large conformational changes in the dengue virus NS2B-NS3 protease: fold analysis by pseudocontact shifts. J Am Chem Soc 133:19205–19215
Fernandez C, Hilty C, Wider G, Guntert P, Wuthrich K (2004) NMR structure of the integral membrane protein OmpX. J Mol Biol 336:1211–1221
Gardner KH, Kay LE (1998) The Use of 2H, 13C, 15N multidimensional NMR to study the structure and dynamics of proteins. Ann Rev of Biophys Biomol Struct 27:357–406
Gautier A, Mott HR, Bostock MJ, Kirkpatrick JP, Nietlispach D (2010) Structure determination of the seven-helix transmembrane receptor sensory rhodopsin II by solution NMR spectroscopy. Nat Struct Mol Biol 17:768–774
Goto NK, Kay LE (2000) New developments in isotope labeling strategies for protein solution NMR spectroscopy. Curr Opin Struct Biol 10:585–592
Graham B et al (2011) DOTA-amide lanthanide tag for reliable generation of pseudocontact shifts in protein NMR spectra. Bioconjug Chem 22:2118–2125
Gront D, Kulp DW, Vernon RM, Strauss CE, Baker D (2011) Generalized fragment picking in Rosetta: design, protocols and applications. PLoS ONE 6:e23294
Hagn F, Nasr ML, Wagner G (2018) Assembly of phospholipid nanodiscs of controlled size for structural studies of membrane proteins by NMR. Nat Protoc 13:79–98
Hiller S et al (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210
Hyberts SG, Arthanari H, Robson SA, Wagner G (2014) Perspectives in magnetic resonance: NMR in the post-FFT era. J Magn Reson 241:60–73
Jaremko M et al (2016) High-resolution NMR determination of the dynamic structure of membrane proteins. Angew Chem Int Ed Engl 55:10518–10521
Jiang WX, Gu XH, Dong X, Tang C (2017) Lanthanoid tagging via an unnatural amino acid for protein structure characterization. J Biomol NMR 67:273–282
Jones DT (1999) Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202
Koehler J, Meiler J (2011) Expanding the utility of NMR restraints with paramagnetic compounds: background and practical aspects. Prog Nucl Magn Reson Spectrosc 59:360–389
Kuenze G, Bonneau R, Leman JK, Meiler J (2019) Integrative protein modeling in RosettaNMR from sparse paramagnetic restraints. Structure 27:1721–1734e5
Lange OF et al (2012) Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci 109:10873–10878
Leman JK, Mueller R, Karakas M, Woetzel N, Meiler J (2013) Simultaneous prediction of protein secondary structure and transmembrane spans. Proteins 81:1127–1140
Lin EC, Opella SJ (2014) Covariance spectroscopy in high-resolution multi-dimensional solid-state NMR. J Magn Reson 239:57–60
Loh CT et al (2013) Lanthanide tags for site-specific ligation to an unnatural amino acid and generation of pseudocontact shifts in proteins. Bioconjug Chem 24:260–268
MacKenzie KR, Prestegard JH, Engelman DM (1997) A transmembrane helix dimer: structure and implications. Science 276:131–133
Miclet E et al (2004) Relaxation-optimized NMR spectroscopy of methylene groups in proteins and nucleic acids. J Am Chem Soc 126:10560–10570
Milles S et al (2012) Click strategies for single-molecule protein fluorescence. J Am Chem Soc 134:5187–5195
Muntener T, Joss D, Haussinger D, Hiller S (2022) Pseudocontact shifts in biomolecular NMR spectroscopy. Chem Rev 122:9422–9467
Nasr ML et al (2017) Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods 14:49–52
Nietlispach D (2005) Suppression of anti-TROSY lines in a sensitivity enhanced gradient selection TROSY scheme. J Biomol NMR 31:161–166
Okwei ENN et al (2022) First crystal structure of a non-canonical amino acid linked to a paramagnetic lanthanide tag facilitates protein structure determination using NMR-derived restraints. bioRxiv (2022).
Orton HW, Huber T, Otting G (2020) Paramagpy: software for fitting magnetic susceptibility tensors using paramagnetic effects measured in NMR spectra. Magnetic Resonance 1:1–12
Otomo T, Ito N, Kyogoku Y, Yamazaki T (1999) NMR observation of selected segments in a larger protein: central-segment isotope labeling through intein-mediated ligation. Biochemistry 38:16040–16044
Oxenoid K, Chou JJ (2005) The structure of phospholamban pentamer reveals a channel-like architecture in membranes. Proc Natl Acad Sci USA 102:10870–10875
Oxenoid K, Kim HJ, Jacob J, Sonnichsen FD, Sanders CR (2004) NMR assignments for a helical 40 kDa membrane protein. J Am Chem Soc 126:5048–5049
Palmer MR et al (2015) Sensitivity of nonuniform sampling NMR. J Phys Chem B 119:6502–6515
Park SH et al (2015) Paramagnetic relaxation enhancement of membrane proteins by incorporation of the metal-chelating unnatural amino acid 2-amino-3- (8-hydroxyquinolin-3-yl)propanoic acid (HQA). J Biomol NMR 61:185–196
Pervushin K, Riek R, Wider G, Wuethrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Romanelli A, Shekhtman A, Cowburn D, Muir TW (2004) Semisynthesis of a segmental isotopically labeled protein splicing precursor: NMR evidence for an unusual peptide bond at the N-extein-intein junction. Proc Natl Acad Sci USA 101:6397–6402
Schmitz C, Vernon R, Otting G, Baker D, Huber T (2012) Protein structure determination from pseudocontact shifts using ROSETTA. J Mol Biol 416:668–677
Seigneuret M, Kainosho M (1993) Localisation of methionine residues in bacteriorhodopsin by carbonyl 13C-NMR with sequence-specific assignments. FEBS Lett 327:7–12
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Simons KT, Kooperberg C, Huang E, Baker D (1997) Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol 268:209–225
Sprangers R, Gribun A, Hwang PM, Houry WA, Kay LE (2005) Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc Natl Acad Sci USA 102:16678–16683
Stanton-Cook M, Su X, Otting G, Huber T (2018) PyParaTools—software for working with paramagnetic NMR data
Suiter CL et al (2014) Sensitivity gains, linearity, and spectral reproducibility in nonuniformly sampled multidimensional MAS NMR spectra of high dynamic range. J Biomol NMR 59:57–73
Susac L, Horst R, Wuthrich K (2014) Solution-NMR characterization of outer-membrane protein A from E. coli in lipid bilayer nanodiscs and detergent micelles. Chembiochem 15:995–1000 (2014).
Tian C et al (2005) Membrane protein preparation for TROSY NMR screening. Methods Enzymol 394:321–334
Trbovic N et al (2005) Efficient strategy for the rapid backbone assignment of membrane proteins. J Am Chem Soc 127:13504–13505
Tugarinov V, Kay LE (2005) Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem 6:1567–1577
Tugarinov V, Hwang PM, Kay LE (2004a) Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu Rev Biochem 73:107–146
Tugarinov V, Sprangers R, Kay LE (2004b) Line narrowing in methyl-TROSY using zero-quantum 1H–13C NMR spectroscopy. J Am Chem Soc 126:4921–4925
Weigelt J, Miles CS, Dixon NE, Otting G (1998) Backbone NMR assignments and secondary structure of the N-terminal domain of DnaB helicase from E. coli. J Biomol NMR 11:233–234 (1998).
Williams CJ et al (2018) MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci 27:293–315
Yagi H, Tsujimoto T, Yamazaki T, Yoshida M, Akutsu H (2004) Conformational change of H+-ATPase beta monomer revealed on segmental isotope labeling NMR spectroscopy. J Am Chem Soc 126:16632–16638
Young TS, Ahmad I, Yin JA, Schultz PG An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol 395:361–74 (2010).
Zhao B, Baisden JT, Zhang Q (2020) Probing excited conformational states of nucleic acids by nitrogen CEST NMR spectroscopy. J Magn Reson 310:106642
Zhou Y et al (2008) NMR solution structure of the integral membrane enzyme DsbB: functional insights into DsbB-catalyzed disulfide bond formation. Mol Cell 31:896–908
Zuger S, Iwai H (2005) Intein-based biosynthetic incorporation of unlabeled protein tags into isotopically labeled proteins for NMR studies. Nat Biotechnol 23:736–740
Acknowledgements
This work was supported by NIH grants R01 GM080403, R01 HL122010, and R01 GM129261. The authors further acknowledge funding by the Deutsche Forschungsgemeinschaft (DFG) through SFB1423, project number 421152132. Jens Meiler is supported by a Humboldt Professorship of the Alexander von Humboldt Foundation. This work was conducted using the resources of the Biomolecular NMR facility and the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University. We especially thank Prof. Markus Voehler for technical assistance with the NMR experiments. We thank Dr. Otting of the Australian National University for providing a test batch of the C3-based paramagnetic tag. All subsequent batches of the C3-based paramagnetic tag were synthesized in-house by Dr. Kwangho Kim of the Vanderbilt Institute of Chemical Biology, Molecular Design and Synthesis Center, Vanderbilt University, Nashville, TN 37232-0412.
Funding
Funding was provided by National Institutes of Health (R01 GM080403) and Deutsche Forschungsgemeinschaft (421152132).
Author information
Authors and Affiliations
Contributions
Conceptualization, KVL, GK, and JM; Data curation, KVL, GK, JRM, EO, KEL, LP, SG and HD; Formal analysis, KVL and GK; Funding acquisition, JM; Investigation, KVL, GK, and JM; Methodology, KVL, GK, and IC; Supervision, KVL and JM; Writing—original draft, KVL; Writing—review and editing, KVL, GK, and JM. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ledwitch, K.V., Künze, G., McKinney, J.R. et al. Sparse pseudocontact shift NMR data obtained from a non-canonical amino acid-linked lanthanide tag improves integral membrane protein structure prediction. J Biomol NMR 77, 69–82 (2023). https://doi.org/10.1007/s10858-023-00412-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-023-00412-9