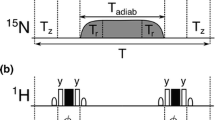
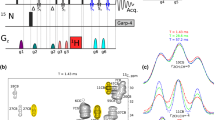
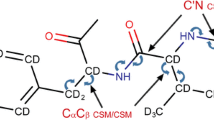
Abstract
The relaxation interference between dipole–dipole interactions of two separate spin pairs carries structural and dynamics information. In particular, when compared to individual dynamic behavior of those spin pairs, such cross-correlated relaxation (CCR) rates report on the correlation between the spin pairs. We have recently mapped out correlated motion along the backbone of the protein GB3, using CCR rates among and between consecutive HN–N and Hα–Cα dipole–dipole interactions. Here, we provide a detailed account of the measurement of the four types of CCR rates. All rates were obtained from at least two different pulse sequences, of which the yet unpublished ones are presented. Detailed comparisons between the different methods and corrections for unwanted pathways demonstrate that the averaged CCR rates are highly accurate and precise with errors of 1.5–3% of the entire value ranges.










Similar content being viewed by others
References
Bonomi M, Camilloni C, Cavalli A, Vendruscolo M (2016) Metainference: a Bayesian inference method for heterogeneous systems. Sci Adv 2:e1501177
Bouvignies G, Bernado P, Meier S, Cho K, Grzesiek S, Brüschweiler R, Blackledge M (2005) Identification of slow correlated motions in proteins using residual dipolar and hydrogen-bond scalar couplings. Proc Natl Acad Sci USA 102:13885–13890
Bremi T, Brüschweiler R (1997) Locally anisotropic internal polypeptide backbone dynamics by NMR relaxation. J Am Chem Soc 119:6672–6673
Bremi T, Brüschweiler R, Ernst RR (1997) A protocol for the interpretation of side-chain dynamics based on NMR relaxation: application to phenylalanines in antamanide. J Am Chem Soc 119:4272–4284
Brüschweiler R, Ernst RR (1992) Molecular dynamics monitored by cross-correlated cross relaxation of spins quantized along orthogonal axes. J Chem Phys 96:1758–1766
Brutscher B, Skrynnikov NR, Bremi T, Brüschweiler R, Ernst RR (1998) Quantitative investigation of dipole-CSA cross-correlated relaxation by ZQ/DQ spectroscopy. J Magn Reson 130:346–351
Carlomagno T, Maurer M, Hennig M, Griesinger C (2000) Ubiquitin backbone motion studied via NHN–C′Cα dipolar–dipolar and C′–C′Cα/NHN CSA–dipolar cross-correlated relaxation. J Am Chem Soc 122:5105–5113
Carlomagno T, Bermel W, Griesinger C (2003) Measuring the χ1 torsion angle in protein by CH–CH cross-correlated relaxation: a new resolution-optimised experiment. J Biomol NMR 27:151–157
Case DA (1999) Calculation of nmr dipolar coupling strengths in modelpeptides. J Biomol NMR 15:95–102
Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skleton NJ (2007) Protein NMR spectroscopy. Principles and pratice. Academic Press, San Diego
Chiarparin E, Pelupessy P, Ghose R, Bodenhausen G (1999) Relaxation of two-spin coherence due to cross-correlated fluctuations of dipole-dipole couplings and anisotropic shifts in nmr of 15N,13C-labeled biomolecules. J Am Chem Soc 121:6876–6883
Chiarparin E, Pelupessy P, Ghose R, Bodenhausen G (2000) Relative orientation of CαHα-bond vectors of successive residues in proteins through cross-correlated relaxation in NMR. J Am Chem Soc 122:1758–1761
Clore GM, Schwieters CD (2006) Concordance of residual dipolar couplings, backbone order parameters and crystallographic B-factors for a small alpha/beta protein: a unified picture of high probability, fast atomic motions in proteins. J Mol Biol 355:879–886
Daragan VA, Mayo KH (1997) Motional model analyses of protein and peptide dynamics using 13C and 15N NMR relaxation. Prog Nucl Magn Reson Spectrosc 31:63–105
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax (1995) nmrPipe—a multidimensional spectral processing system based on unix pipes. J Biomol NMR 6:277–293
Fenwick RB, Esteban-Martin S, Richter B, Lee D, Walter KFA, Milanovic D, Becker S, Lakomek NA, Griesinger C, Salvatella X (2011) Weak long-range correlated motions in a surface patch of ubiquitin involved in molecular recognition. J Am Chem Soc 133:10336–10339
Fenwick RB, Schwieters CD, Vögeli B (2016) Direct investigation of slow correlated dynamics in proteins via dipolar interactions. J Am Chem Soc 138:8412–8421
Früh D, Tolman JR, Bodenhausen G, Zwahlen C (2001) Cross-correlated chemical shift modulation: a signature of slow internal motions in proteins. J Am Chem Soc 123:4810–4816
Geen H, Freeman R (1991) Band-selective radiofrequency pulses. J Magn Reson 93:93–141
Goldman M (1984) Interference effects in the relaxation of a pair of unlike spin-1/2 nuclei. J Magn Reson 60:437–452
Hall JB, Fushman D (2003) Characterization of the overall and local dynamics of a protein with intermediate rotational anisotropy: differentiating between conformational exchange and anisotropic diffusion in the B3 domain of protein G. J Biomol NMR 27:261–275
Hubbard PS (1958) Nuclear magnetic relaxation of three and four spin molecules in a liquid. Phys Rev 109:1153–1158
Johnson BA, Blevins RA (1994) NMR view: a computer program for the visualization and analysis of NMR data. J Biomol NMR 5:603–614
Kay LE (2005) NMR studies of protein structure and dynamics. J Magn Reson 173:193–207
Kay LE, Keifer P, Saarinen T (1992) Pure absorption gradient enhanced heteronuclear single quantum correlation spectroscopy with improved sensitivity. J Am Chem Soc 114:10663–10665
Kleckner IR, Foster MP (2011) An introduction to NMR-based approaches for measuring protein dynamics. Biochim Biophys Acta 1814:942–968
Kloiber K, Schuler W, Konrat R (2002) Automated NMR determination of protein backbone dihedral angles from cross-correlated spin relaxation. J Biomol NMR 22:349–363
Korzhnev DM, Billeter M, Arseniev AS, Orekhov VY (2001) Nmr studies of brownian tumbling and internal motions in proteins. Prog Nucl Magn Reson Spectrosc 38:197–266
Kumar A, Grace CRR, Madhu PK (2000) Cross-correlations in NMR. Prog Nucl Magn Reson Spectrosc 37:191–319
Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M (2005) Simultaneous determination of protein structure and dynamics. Nature 433:128–132
Lipari G, Szabo A (1982a) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules 1. Theory and range of validity. J Am Chem Soc 104:4546–4559
Lipari G, Szabo A (1982b) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc 104:4559–4570
Lundström P, Mulder FAA, Akke M (2005) Correlated dynamics of consecutive residues reveal transient and cooperative unfolding of secondary structure in proteins. Proc Natl Acad Sci USA 102:16984–16989
Marion D, Ikura M, Tschudin R, Bax A (1989) Rapid recording of 2D nmr spectra without phase cycling. Application to study of hydrogen exchange in proteins. J Magn Reson 85:393–399
Markwick PRL, Bouvignies G, Blackledge M (2007) Exploring multiple timescale motions in protein GB3 using accelerated molecular dynamics and nmr spectroscopy. J Am Chem Soc 129:4724–4730
McConnell HM (1956) Effect of anisotropic hyperfine interactions on paramagnetic relaxation in liquids. J Chem Phys 25:709–711
Mori M, Kateb F, Bodenhausen G, Piccioli M, Abergel D (2010) Toward structural dynamics: protein motions viewed by chemical shift modulations and direct detection of C′N multiple-quantum relaxation. J Am Chem Soc 132:3594–3600
Olsson S, Vögeli B, Cavalli A, Boomsma W, Ferkinghoff-Borg J, Lindorff-Larsen K, Hamelryck T (2014) Probabilistic determination of native state ensembles of proteins. J Chem Theory Comput 10:3484–3491
Palmer AG (2016) A dynamic look backward and forward. J Magn Reson 266:73–80
Pellecchia M, Pang Y, Wang L, Kurochkin AV, Kumar A, Zuiderweg ERP (1999) Quantitative measurement of cross-correlations between 15N and 13CO chemical shift anisotropy relaxation mechanisms by multiple quantum NMR. J Am Chem Soc 121:9165–9170
Pelupessy P, Chiarparin E, Ghose R, Bodenhausen G (1999a) Efficient determination of angles subtended by Cα–Hα and N–HN vectors in proteins via dipole-dipole cross-correlation. J Biomol NMR 13:375–380
Pelupessy P, Chiarparin E, Ghose R, Bodenhausen G (1999b) Simultaneous determination of Phi and Psi angles in proteins from measurements of cross-correlated relaxation effects. J Biomol NMR 14:277–280
Pelupessy P, Ravindranathan S, Bodenhausen G (2003) Correlated motions of successive amide N–H bonds in proteins. J Biomol NMR 25:265–280
Reif B, Hennig M, Griesinger C (1997) Direct measurement of angles between bond vectors in high-resolution NMR. Science 276:1230–1233
Reif B, Diener A, Hennig M, Maurer M, Griesinger C (2000) Cross-correlated relaxation for the measurement of angles between tensorial interactions. J Magn Reson 143:45–68
Schwalbe H, Carlomagno T, Hennig M, Junker J, Reif B, Richter C, Griesinger C (2001) Cross-correlated relaxation for measurement of angles between tensorial interactions. Methods Enzymol 338:35–81
Shaka AJ, Keeler J, Frenkiel T, Freeman R (1983) An improved sequence for broadband decoupling: WALTZ-16. J Magn Reson 52:335–338
Shaka AJ, Barker PB, Freeman R (1985) Computer-optimized decoupling scheme for wideband application and low-level operation. J Magn Reson 64:547–552
Shapiro YE (2013) NMR spectroscopy on domain dynamics in biomacromolecules. Prog Biophys Mol Biol 112:58–117
Skrynnikov NR, Konrat R, Muhandiram DR, Kay LE (2000) Relative orientation of peptide planes in proteins is reflected in carbonyl-carbonyl chemical shift anisotropy cross-correlated spin relaxation. J Am Chem Soc 122:7059–7071
Takahashi H, Shimada I (2007) Pairwise NMR experiments for the determination of protein backbone dihedral angle Phi based on cross-correlated spin relaxation. J Biomol NMR 37:179–185
Tjandra N, Feller SE, Pastor RW, Bax A (1995) Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. J Am Chem Soc 117:12562–12566
Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH (2001) Structural and dynamic analysis of residual dipolar coupling data for proteins. J Am Chem Soc 123:1416–1424
Torchia D (2015) NMR studies of dynamic biomolecular conformational ensembles. Prog Nucl Magn Reson Spectrosc 84–85:14–32
Ulmer TS, Ramirez BE, Delaglio F, Bax A (2003) Evaluation of backbone proton positions and dynamics in a small protein by liquid crystal NMR spectroscopy. J Am Chem Soc 125:9179–9191
Vammi V, Song G (2015) Ensembles of a small number of conformations with relative populations. J Biomol NMR 63:341–351
Vögeli B (2010) Comprehensive description of NMR cross-correlated relaxation under anisotropic molecular tumbling and correlated local dynamics on all time scales. J Chem Phys 133:014501
Vögeli B (2011) How uniform is the peptide plane geometry? A high-accuracy NMR study of dipolar Cα–C′/H N–N cross-correlated relaxation. J Biomol NMR 50:315–329
Vögeli B (2013) Full relaxation matrix analysis of apparent cross-correlated relaxation rates in four-spin systems. J Magn Reson 226:52–63
Vögeli B, Pervushin K (2002) Trosy experiment for refinement of backbone ψ and ϕ by simultaneous measurements of cross-correlated relaxation rates and 3,4JHaHN coupling constants. J Biomol NMR 24:291–300
Vögeli B, Riek R (2010) Side chain: backbone projections in aromatic and ASX residues from NMR cross-correlated relaxation. J Biomol NMR 46:135–147
Vögeli B, Yao LS (2009) Correlated dynamics between protein HN and HC bonds observed by NMR cross relaxation. J Am Chem Soc 131:3668–3678
Vögeli B, Ying JF, Grishaev A, Bax A (2007) Limits on variations in protein backbone dynamics from precise measurements of scalar couplings. J Am Chem Soc 129:9377–9385
Vögeli B, Yao L, Bax A (2008) Protein backbone motions viewed by intraresidue and sequential HN–Hα residual dipolar couplings. J Biomol NMR 41:17–28
Vögeli B, Friedmann M, Leitz D, Sobol A, Riek R (2010) Quantitative determination of NOE rates in perdeuterated and protonated proteins: practical and theoretical aspects. J Magn Reson 204:290–302
Vögeli B, Olsson S, Riek R, Güntert P (2015) Compiled data set of exact NOE distance limits, residual dipolar couplings and scalar couplings for the protein GB3. Data in Brief 50:99–106
Vögeli B, Olsson S, Güntert P, Riek R (2016) The exact NOE as an alternative in ensemble structure determination. Biophys J 110:113–126
Vugmeyster L, McKnight CJ (2008) Slow motions in chicken villin headpiece subdomain probed by cross-correlated NMR relaxation of amide NH bonds in successive residues. Biophys J 95:5941–5950
Vugmeyster L, McKnight CJ (2009) Phosphorylation-induced changes in backbone dynamics of the dematin headpiece C-terminal domain. J Biomol NMR 43:39–50
Vugmeyster L, Pelupessy P, Vugmeister BE, Abergel D, Bodenhausen G (2004) Cross-correlated relaxation in NMR of macromolecules in the presence of fast and slow internal dynamics. CR Physique 5: 377–386
Werbelow LG, Marshall AG (1973) Internal rotation and nonexponential methyl nuclear relaxation for macromolecules. J Magn Reson 11:299–313
Yang DW, Kay LE (1998) Determination of the protein backbone dihedral angle ψ from a combination of NMR-derived cross-correlation spin relaxation rates. J Am Chem Soc 120:9880–9887
Yang D, Konrat R, Kay LE (1997) A sensitive pulse scheme for measuring the backbone dihedral angle ψ based on cross-correlation between 1Hα–13Cα dipolar and 13C′ (carbonyl) chemical shift anisotropy relaxation interactions. J Am Chem Soc 119:11938–11940
Yao L, Vögeli B, Torchia DA, Bax A (2008a) Simultaneous NMR study of protein structure and dynamics using conservative mutagenesis. J Phys Chem B 112:6045–6056
Yao L, Vögeli B, Ying JF, Bax A (2008b) Nmr determination of amide N–H equilibrium bond length from concerted dipolar coupling measurements. J Am Chem Soc 130:16518–16520
Acknowledgements
This work was supported by a start-up package at the University of Colorado Denver and the Swiss National Science Foundation with Grant 140214 to B.V.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vögeli, B. Cross-correlated relaxation rates between protein backbone H–X dipolar interactions. J Biomol NMR 67, 211–232 (2017). https://doi.org/10.1007/s10858-017-0098-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-017-0098-5