Abstract
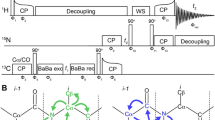
This study demonstrates a novel spectral editing technique for protein solid-state NMR (SSNMR) to simplify the spectrum drastically and to reduce the ambiguity for protein main-chain signal assignments in fast magic-angle-spinning (MAS) conditions at a wide frequency range of 40–80 kHz. The approach termed HIGHLIGHT (Wang et al., in Chem Comm 51:15055–15058, 2015) combines the reverse 13C, 15N-isotope labeling strategy and selective signal quenching using the frequency-selective REDOR pulse sequence under fast MAS. The scheme allows one to selectively observe the signals of “highlighted” labeled amino-acid residues that precede or follow unlabeled residues through selectively quenching 13CO or 15N signals for a pair of consecutively labeled residues by recoupling 13CO–15N dipolar couplings. Our numerical simulation results showed that the scheme yielded only ~15 % loss of signals for the highlighted residues while quenching as much as ~90 % of signals for non-highlighted residues. For lysine-reverse-labeled micro-crystalline GB1 protein, the 2D 15N/13Cα correlation and 2D 13Cα/13CO correlation SSNMR spectra by the HIGHLIGHT approach yielded signals only for six residues following and preceding the unlabeled lysine residues, respectively. The experimental dephasing curves agreed reasonably well with the corresponding simulation results for highlighted and quenched residues at spinning speeds of 40 and 60 kHz. The compatibility of the HIGHLIGHT approach with fast MAS allows for sensitivity enhancement by paramagnetic assisted data collection (PACC) and 1H detection. We also discuss how the HIGHLIGHT approach facilitates signal assignments using 13C-detected 3D SSNMR by demonstrating full sequential assignments of lysine-reverse-labeled micro-crystalline GB1 protein (~300 nmol), for which data collection required only 11 h. The HIGHLIGHT approach offers valuable means of signal assignments especially for larger proteins through reducing the number of resonance and clarifying multiple starting points in sequential assignment with enhanced sensitivity.






Similar content being viewed by others
References
Ahuja S et al (2013) A model of the membrane-bound cytochrome b(5)-cytochrome P450 complex from NMR and mutagenesis data. J Biol Chem 288:22080–22095
Akinlolu RD, Nam M, Qiang W (2015) Competition between fibrillation and induction of vesicle fusion for the membrane-associated 40-residue beta-amyloid peptides. Biochemistry 54:3416–3419
Andronesi OC, von Bergen M, Biernat J, Seidel K, Griesinger C, Mandelkow E, Baldus M (2008) Characterization of Alzheimer’s-like paired helical filaments from the core domain of tau protein using solid-state NMR spectroscopy. J Am Chem Soc 130:5922–5928
Bak M, Nielsen NC (1997) REPULSION, a novel approach to efficient powder averaging in solid-state NMR. J Magn Reson 125:132–139
Banigan JR, Gayen A, Traaseth NJ (2013) Combination of N-15 reverse labeling and afterglow spectroscopy for assigning membrane protein spectra by magic-angle-spinning solid-state NMR: application to the multidrug resistance protein EmrE. J Biomol NMR 55:391–399
Bhate MP, Wylie BJ, Tian L, McDermott AE (2010) Conformational dynamics in the selectivity filter of KcsA in response to potassium ion concentration. J Mol Biol 401:155–166
Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M (2010) Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nature 463:689–692
Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y (2007) Evidence of fibril-like beta-sheet structures in a neurotoxic amyloid intermediate of Alzheimer’s beta-amyloid. Nat Struct Mol Biol 14:1157–1164
Coin I, Beyermann M, Bienert M (2007) Solid-phase peptide synthesis: from standard procedures to the synthesis of difficult sequences. Nat Protoc 2:3247–3256
Franks WT et al (2005) Magic-angle spinning solid-state NMR spectroscopy of the beta 1 immunoglobulin binding domain of protein G (GB1): N-15 and C-13 chemical shift assignments and conformational analysis. J Am Chem Soc 127:12291–12305
Fung BM, Khitrin AK, Ermolaev K (2000) An improved broadband decoupling sequence for liquid crystals and solids. J Magn Reson 142:97–101
Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M (2005) Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc Natl Acad Sci USA 102:15871–15876
Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP (2008) Molecular conformation and dynamics of the Y145Stop variant of human prion protein. Proc Natl Acad Sci USA 105:6284–6289
Hiller M, Higman VA, Jehle S, van Rossum BJ, Kuhlbrandt W, Oschkinat H (2008) 2,3-C-13-labeling of aromatic residues-getting a head start in the magic-angle-spinning NMR assignment of membrane proteins. J Am Chem Soc 130:408
Hong M, Jakes K (1999) Selective and extensive C-13 labeling of a membrane protein for solid-state NMR investigations. J Biomol NMR 14:71–74
Igumenova TI, McDermott AE, Zilm KW, Martin RW, Paulson EK, Wand AJ (2004) Assignments of carbon NMR resonances for microcrystalline ubiquitin. J Am Chem Soc 126:6720–6727
Ishii Y (2001) C-13-C-13 dipolar recoupling under very fast magic angle spinning in solid-state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary-structure determination. J Chem Phys 114:8473–8483
Ishii Y, Tycko R (2000) Sensitivity enhancement in solid state 15 N NMR by indirect detection with high-speed magic angle spinning. J Magn Reson 142:199–204
Ishii Y, Yesinowski JP, Tycko R (2001) Sensitivity enhancement in solid-state C-13 NMR of synthetic polymers and biopolymers by H-1 NMR detection with high-speed magic angle spinning. J Am Chem Soc 123:2921–2922
Jaroniec CP, Tounge BA, Rienstra CM, Herzfeld J, Griffin RG (2000) Recoupling of heteronuclear dipolar interactions with rotational-echo double-resonance at high magic-angle spinning frequencies. J Magn Reson 146:132–139
Kent SBH (1988) Chemical synthesis of peptides and proteins. Annu Rev Biochem 57:957–989
Kigawa T, Muto Y, Yokoyama S (1995) Cell-free synthesis and amino acid-selective stable-isotope labeling of proteins for NMR analysis. J Biomol NMR 6:129–134
Knight MJ et al (2012) Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR. Proc Natl Acad Sci USA 109:11095–11100
Lai J et al (2011) X-ray and NMR crystallography in an enzyme active site: the indoline quinonoid intermediate in tryptophan synthase. J Am Chem Soc 133:4–7
Lange A, Giller K, Hornig S, Martin-Eauclaire MF, Pongs O, Becker S, Baldus M (2006) Toxin-induced conformational changes in a potassium channel revealed by solid-state NMR. Nature 440:959–962
Lewandowski JR, van der Wel PCA, Rigney M, Grigorieff N, Griffin RG (2011) Structural complexity of a composite amyloid fibril. J Am Chem Soc 133:14686–14698
Lizak MJ, Gullion T, Conradi MS (1991) Measurement of like-spin dipole couplings. J Magn Reson 91:254–260
Lopez del Amo JM, Schmidt M, Fink U, Dasari M, Faendrich M, Reif B (2012) An asymmetric dimer as the basic subunit in alzheimer’s disease amyloid beta fibrils. Angew Chem Int Ed 51:6136–6139
Mahalakshmi R, Marassi FM (2008) Orientation of the Escherichia coli outer membrane protein OmpX in phospholipid bilayer membranes determined by solid-state NMR. Biochemistry 47:6531–6538
McDermott A (2009) Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Ann Rev Biophys 38:385–403
Morcombe CR, Zilm KW (2003) Chemical shift referencing in MAS solid state NMR. J Magn Reson 162:479–486
Mueller LJ, Dunn MF (2013) NMR crystallography of enzyme active sites: probing chemically detailed. Three-dimensional structure in tryptophan synthase. Acc Chem Res 46:2008–2017
Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R (2009) Seeded growth of beta-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure. Proc Natl Acad Sci USA 106:7443–7448
Park SH et al (2012) Structure of the chemokine receptor CXCR1 in phospholipid bilayers. Nature 491:779
Parthasarathy S, Nishiyama Y, Ishii Y (2013) Sensitivity and resolution enhanced solid-state NMR for paramagnetic systems and biomolecules under very fast magic angle spinning. Acc Chem Res 46:2127–2135
Petkova AT, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R (2002) A structural model for Alzheimer’s beta-amyloid fibrils based on experimental constraints from solid state NMR. Proc Natl Acad Sci USA 99:16742–16747
Qiang W, Sun Y, Weliky DP (2009) A strong correlation between fusogenicity and membrane insertion depth of the HIV fusion peptide. Proc Natl Acad Sci USA 106:15314–15319
Richter L et al (2010) Amyloid beta 42 peptide (A beta 42)-lowering compounds directly bind to A beta and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proc Natl Acad Sci USA 107:14597–14602
Schmidt-Rohr K, Fritzsching KJ, Liao SY, Hong M (2012) Spectral editing of two-dimensional magic-angle-spinning solid-state NMR spectra for protein resonance assignment and structure determination. J Biomol NMR 54:343–353
Schuetz A et al (2010) Protocols for the sequential solid-state NMR spectroscopic assignment of a uniformly labeled 25 kDa protein: HET-s(1-227). ChemBioChem 11:1543–1551
Shahid SA, Markovic S, Linke D, van Rossum B-J (2012) Assignment and secondary structure of the YadA membrane protein by solid-state MAS NMR. Sci Rep 2
Shi LC, Ahmed MAM, Zhang WR, Whited G, Brown LS, Ladizhansky V (2009) Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump-structural insights. J Mol Biol 386:1078–1093
Sivanandam VN, Jayaraman M, Hoop CL, Kodali R, Wetzel R, van der Wel PCA (2011) The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J Am Chem Soc 133:4558–4566
Staunton D, Schlinkert R, Zanetti G, Colebrook SA, Campbell LD (2006) Cell-free expression and selective isotope labelling in protein NMR. Magn Reson Chem 44:S2–S9
Sun Y, Weliky DP (2009) C-13-C-13 correlation spectroscopy of membrane-associated influenza virus fusion peptide strongly supports a helix-turn-helix motif and two turn conformations. J Am Chem Soc 131:13228–13229
Traaseth NJ, Veglia G (2011) Frequency-selective heteronuclear dephasing and selective carbonyl labeling to deconvolute crowded spectra of membrane proteins by magic angle spinning NMR. J Magn Reson 211:18–24
Verardi R, Shi L, Traaseth NJ, Walsh N, Veglia G (2011) Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc Natl Acad Sci USA 108:9101–9106
Veshtort M, Griffin RG (2006) SPINEVOLUTION: a powerful tool for the simulation of solid and liquid state NMR experiments. J Magn Reson 178:248–282
Vuister GW, Kim SJ, Wu C, Bax A (1994) 2D and 3D NMR-study of phenylalanine residues in proteins by reverse isotopic labeling. J Am Chem Soc 116:9206–9210
Wang S, Ishii Y (2012) Revealing protein structures in solid-phase peptide synthesis by C-13 solid-state NMR: evidence of excessive misfolding for Alzheimer’s beta. J Am Chem Soc 134:2848–2851
Wang S et al (2015) Nano-mole scale sequential signal assignment by 1H-detected protein solid-state NMR. Chem Commun 51:15055–15058
Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, Meier BH (2008) Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science 319:1523–1526
Wickramasinghe NP et al (2009) Nanomole-scale protein solid-state NMR by breaking intrinsic H-1 T-1 boundaries. Nat Methods 6:215–218
Xiao YL et al (2015) A beta(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol 22:499–505
Yamazaki T et al (1998) Segmental isotope labeling for protein NMR using peptide splicing. J Am Chem Soc 120:5591–5592
Zhang Y, Doherty T, Li J, Lu WY, Barinka C, Lubkowski J, Hong M (2010) Resonance assignment and three-dimensional structure determination of a human alpha-defensin, HNP-1, by solid-state NMR. J Mol Biol 397:408–422
Acknowledgments
This study was supported primarily from the U.S. National Science Foundation (CHE 957793 and CHE 1310363) for YI. The instrumentation of the 750 MHz SSNMR at UIC was supported by an NIH HEI Grant (1S10 RR025105).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, S., Matsuda, I., Long, F. et al. Spectral editing at ultra-fast magic-angle-spinning in solid-state NMR: facilitating protein sequential signal assignment by HIGHLIGHT approach. J Biomol NMR 64, 131–141 (2016). https://doi.org/10.1007/s10858-016-0014-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-016-0014-4