Abstract
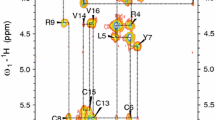
A tendency to dimerize in the presence of lipids was found for the protegrin. The dimer formation by the protegrin-1 (PG-1) is the first step for further oligomeric membrane pore formation. Generally there are two distinct model of PG-1 dimerization in either a parallel or antiparallel β-sheet. But despite the wealth of data available today, protegrin dimer structure and pore formation is still not completely understood. In order to investigate a more detailed dimerization process of PG-1 and if it will be the same for another type of protegrins, in this work we used a high-resolution NMR spectroscopy for structure determination of protegrin-3 (RGGGL-CYCRR-RFCVC-VGR) in the presence of perdeuterated DPC micelles and demonstrate that PG-3 forms an antiparallel NCCN dimer with a possible association of these dimers. This structural study complements previously published solution, solid state and computational studies of PG-1 in various environments and validate the potential of mean force simulations of PG-1 dimers and association of dimers to form octameric or decameric β-barrels.





Similar content being viewed by others
References
Afonin S, Grage SL, Ieronimo M, Wadhwani P, Ulrich AS (2011) Temperaturedependent transmembrane insertion of the amphiphilic peptide PGLa in lipid bilayers observed by solid state 19F NMR spectroscopy. J Am Chem Soc 130:16512–16514. doi:10.1021/ja803156d
Arora A, Tamm LK (2001) Biophysical approaches to membrane protein structure determination. Curr Opin Struct Biol 11:540–547. doi:10.1016/S0959-440X(00)00246-3
Blochin DS et al (2013) Spatial structure of heptapeptide Glu-Ile-Leu-Asn-His-Met-Lys, a fragment of the HIV enhancer prostatic acid phosphatase, in aqueous and SDS micelle solutions. J Mol Struct 1033:59–66. doi:10.1016/j.molstruc.2012.08.018
Buffy JJ, Hong T, Yamaguchi S, Waring AJ, Lehrer RI, Hong M (2003) Solid-state NMR investigation of the depth of insertion of protegrin-1 in lipid bilayers using paramagnetic Mn2+. Biophys J 85:2363–2373. doi:10.1016/S0006-3495(03)74660-8
Chan DI, Prenner EJ, Vogel HJ (2006) Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta 1758:1184–1202. doi:10.1016/j.bbamem.2006.04.006
Chen VB et al (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D 66:12–21. doi:10.1107/S0907444909042073
Choi MK et al (2014) Defining the genetic relationship of protegrin-related sequences and the in vivo expression of protegrins. FEBS J 281:5420–5431. doi:10.1111/Febs.13072
Cho Y, Turner JS, Dinh N-N, Lehrer RI (1998) Activity of protegrins against yeast-phase candida albicans. Infect Immun 66(6):2486–2493
Davis IW et al (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi:10.1093/Nar/Gkm216
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) Nmrpipe—a multidimensional spectral processing system based on unix pipes. J Biomol NMR 6:277–293. doi:10.1007/Bf00197809
Elad S, Epstein JB, Raber-Durlacher J, Donnelly P, Strahilevitz J (2012) The antimicrobial effect of Iseganan HCl oral solution in patients receiving stomatotoxic chemotherapy: analysis from a multicenter, double-blind, placebo-controlled, randomized, phase III clinical trial. J Oral Pathol Med 41:229–234. doi:10.1111/j.1600-0714.2011.01094.x
Epand RF, Ramamoorthy A, Epand RM (2006) Membrane lipid composition and the interaction with Pardaxin: the role of cholesterol. Protein Pept Lett 13:1–5. doi:10.2174/0929866510602010001
Fahrner RL, Dieckmann T, Harwig SSL, Lehrer RI, Eisenberg D, Feigon J (1996) Solution structure of protegrin-1, a broad-spectrum antimicrobial peptide from porcine leukocytes. Chem Biol 3:543–550. doi:10.1016/S1074-5521(96)90145-3
Galiullina LF, Blokhin DS, Aganov AV, Klochkov VV (2012) Investigation of cholesterol + model of biological membrane complex by NMR spectroscopy. Magn Reson Solids 14:12204–12210
Gottler LM, La De, Salud-Bea R, Shelburne CE, Ramamoorthy A, Marsh ENG (2008) Using fluorous amino acids to probe the effects of changing hydrophobicity on the physical and biological properties of the -hairpin antimicrobial peptide protegrin-1. Biochemistry 47:9243–9250. doi:10.1021/bi801045n
Haney EF, Hunter HN, Matsuzaki K, Vogel HJ (2009) Solution NMR studies of amphibian antimicrobial peptides: linking structure to function? Biochim Biophys Acta 1788:1639–1655. doi:10.1016/j.bbamem.2009.01.002
Hiller S, Wagner G (2009) The role of solution NMR in the structure determinations of VDAC-1 and other membrane proteins. Curr Opin Struct Biol 19:396–401. doi:10.1016/j.sbi.2009.07.013
Hong M (2006) Solid-state NMR studies of the structure, dynamics, and assembly of beta-sheet membrane peptides and alpha-helical membrane proteins with antibiotic activities. Acc Chem Res 39:176–183. doi:10.1021/ar040037e
Hwang TL, Shaka AJ (1995) Water suppression that works—excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson Ser A 112:275–279. doi:10.1006/jmra.1995.1047
Jang H, Ma B, Woolf TB, Nussinov R (2006) Interaction of protegrin-1 with lipid bilayers: membrane thinning effect. Biophys J 91:2848–2859. doi:10.1529/biophysj.106.084046
Jang H, Ma BY, Nussinov R (2007) Conformational study of the protegrin-I (PG-I) dimer interaction with lipid bilayers and its effect. BMC Struct Biol 7:21. doi:10.1186/1472-6807-7-21
Jang H, Ma B, Lal R, Nussinov R (2008) Models of toxic beta-sheet channels of protegrin-1 suggest a common subunit organization motif shared with toxic alzheimer beta-amyloid ion channels. Biophys J 95:4631–4642. doi:10.1529/biophysj.108.134551
Jenssen H, Hamill P, Hancock REW (2006) Peptide antimicrobial agents. Clin Microbiol Rev 19:491–511. doi:10.1128/Cmr.00056-05
Khayrutdinov BI, Bae WJ, Yun YM, Lee JH, Tsuyama T, Kim JJ, Hwang E, Ryu KS, Cheong HK, Cheong C, Ko JS, Enomoto T, Karplus PA, Güntert P, Tada S, Jeon YH, Cho Y (2009) Structure of the Cdt1 C-terminal domain: conservation of the winged helix fold in replication licensing factors. Protein Sci 18:2252–2264. doi:10.1002/pro.236
Kim K, Khayrutdinov BI, Lee CK, Cheong HK, Kang SW, Park H, Lee S, Kim YG, Jee JG, Rich, Kim KK, Jeon YH (2011) Solution structure of the Zβ domain of human DNA-dependent activator of IFN-regulatory factors and its binding modes to B-and Z-DNAs. Proc Natl Acad Sci USA 108:6921–6926. doi:10.1073/pnas.1014898107
Kokryakov VN et al (1993) Protegrins—leukocyte antimicrobial peptides that combine features of corticostatic defensins and tachyplesins. FEBS Lett 327:231–236. doi:10.1016/0014-5793(93)80175-T
Langham AA, Ahmad AS, Kaznessis YN (2008) On the nature of antimicrobial activity: a model for protegrin-1 pores. J Am Chem Soc 130:4338–4346. doi:10.1021/Ja0780380
Lazaridis T, He Y, Prieto L (2013) Membrane interactions and pore formation by the antimicrobial peptide protegrin. Biophys J 104:633–642. doi:10.1016/j.bpj.2012.12.038
Loury DJ, Embree JR, Steinberg DA, Sonis ST, Fiddes JC (1999) Effect of local application of the antimicrobial peptide IB-367 on the incidence and severity of oral mucositis in hamsters. Oral Surg Oral Med O 87:544–551. doi:10.1016/S1079-2104(99)70131-9
Mani R, Tang M, Wu X, Buffy JJ, Waring AJ, Sherman MA, Hong M (2006) Membrane-bound dimer structure of a beta-hairpin antimicrobial peptide from rotational-echo double-resonance solid-state NMR. Biochemistry 45:8341–8349. doi:10.1021/Bi060305b
Ovchinnikova TV, Shenkarev ZO, Balandin SV, Nadezhdin KD, Paramonov AS, Kokryakov VN, Arseniev AS (2008) Molecular insight into mechanism of antimicrobial action of the beta-hairpin peptide arenicin: specific oligomerization in detergent micelles. Biopolymers 89:455–464. doi:10.1002/bip.20865
Papo N, Shai Y (2004) Effect of drastic sequence alteration and D-amino acid incorporation on the membrane binding behavior of lytic peptides. Biochemistry 43:6393–6403. doi:10.1021/bi049944h
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi:10.1002/Jcc.20084
Piotto M, Saudek V, Sklenar V (1992) Gradient-tailored excitation for single-quantum NMR-spectroscopy of aqueous-solutions. J Biomol NMR 2:661–665. doi:10.1007/Bf02192855
Porcelli F, Verardi R, Shi L, Henzler-Wildman KA, Ramamoorthy A, Veglia G (2008) NMR structure of the cathelicidin-derived human antimicrobial peptide LL-37 in dodecylphosphocholine micelles. Biochemistry 47:5565–5572. doi:10.1021/bi702036s
Powers JP, Tan A, Ramamoorthy A, Hancock RE (2005) Solution structure and interaction of the antimicrobial polyphemusins with lipid membranes. Biochemistry 44:15504–15513. doi:10.1021/bi051302m
Ramamoorthy A (2009) Beyond NMR spectra of antimicrobial peptides: dynamical images at atomic resolution and functional insights. Solid State Nucl Magn Reson 35:201–207. doi:10.1016/j.ssnmr.2009.03.003
Ramamoorthy A, Lee DK, Santos JS, Henzler-Wildman KA (2008) Nitrogen-14 solid-state NMR spectroscopy of aligned phospholipid bilayers to probe peptide–lipid interaction and oligomerization of membrane associated peptides. J Am Chem Soc 130:11023–11029. doi:10.1021/ja802210u
Roumestand C, Louis V, Aumelas A, Grassy G, Calas B, Chavanieu A (1998) Oligomerization of protegrin-1 in the presence of DPC micelles. A proton high-resolution NMR study. FEBS Lett 421:263–267. doi:10.1016/S0014-5793(97)01579-2
Saravanan R, Bhattacharjya S (2011) Oligomeric structure of a cathelicidin antimicrobial peptide in dodecylphosphocholine micelle determined by NMR spectroscopy. Biochim Biophys Acta 1808:369–381. doi:10.1016/j.bbamem.2010.10.001
Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package. J Magn Reson 160:65–73. doi:10.1016/S1090-7807(02)00014-9
Shai Y (2002) From innate immunity to de-novo designed antimicrobial peptides. Curr Pharm Des 8:715–725. doi:10.2174/1381612023395367
Sklenar V, Piotto M, Leppik R, Saudek V (1993) Gradient-tailored water suppression for H-1-N-15 HSQC experiments optimized to retain full sensitivity. J Magn Reson Ser A 102:241–245. doi:10.1006/jmra.1993.1098
Syvitski RT, Burton I, Mattatall NR, Douglas SE, Jakeman DL (2005) Structural characterization of the antimicrobial peptide pleurocidin from winter flounder. Biochemistry 44:7282–7293. doi:10.1021/bi0504005
Trotti A et al (2004) A multinational, randomized phase III trial of iseganan HCL oral solution for reducing the severity of oral mucositis in patients receiving radiotherapy for head-and-neck malignancy. Int J Radiat Oncol 58:674–681. doi:10.1016/S0630-3016(04)01627-4
Usachev KS, Filippov AV, Antzutkin ON, Klochkov VV (2013) Use of a combination of the RDC method and NOESY NMR spectroscopy to determine the structure of Alzheimer’s amyloid A beta(10-35) peptide in solution and in SDS micelles. Eur Biophys J Biophy 42:803–810. doi:10.1007/s00249-013-0928-7
Usachev KS, Efimov SV, Kolosova OA, Filippov AV, Klochkov VV (2014a) High-resolution NMR structure of the antimicrobial peptide protegrin-2 in the presence of DPC micelles. J Biomol NMR (in press). doi:10.1007/s10858-014-9885-4
Usachev KS, Filippov AV, Khairutdinov BI, Antzutkin ON, Klochkov VV (2014b) NMR structure of the Arctic mutation of the Alzheimer’s A beta(1-40) peptide docked to SDS micelles. J Mol Struct 1076:518–523. doi:10.1016/j.molstruc.2014.08.030
Vivcharuk V, Kaznessis YN (2010) Dimerization of protegrin-1 in different environments. Int J Mol Sci 11:3177–3194. doi:10.3390/Ijms11093177
Wang G (2007) Determination of solution structure and lipid micelle location of an engineered membrane peptide by using one NMR experiment and one sample. Biochim Biophys Acta 1768:3271–3281. doi:10.1016/j.bbamem.2007.08.005
Yamaguchi S, Hong T, Waring A, Lehrer RI, Hong M (2002) Solid-state NMR investigations of peptide-lipid interaction and orientation of a ss-sheet antimicrobial peptide, protegrin. Biochemistry-US 41:9852–9862. doi:10.1021/Bi0257991
Zamoon J, Nitu F, Karim C, Thomas DD, Veglia G (2005) Mapping the interaction surface of a membrane protein: unveiling the conformational switch of phospholamban in calcium pump regulation. Proc Natl Acad Sci USA 102:4747–4752. doi:10.1073/pnas.0406039102
Zanetti M, Gennaro R, Romeo D (1995) Cathelicidins: a novel protein family with a common proregion and a variable C-terminal antimicrobial domain. FEBS Lett 374:1–5. doi:10.1016/0014-5793(95)01050-O
Zhao CQ, Liu LD, Lehrer RI (1994) Identification of a new member of the protegrin family by cdna cloning. FEBS Lett 346:285–288. doi:10.1016/0014-5793(94)00493-5
Zhao CQ, Ganz T, Lehrer RI (1995) The structure of porcine protegrin genes. FEBS Lett 368:197–202. doi:10.1016/0014-5793(95)00633-K
Acknowledgments
We thank Dr. Andrey Filippov for peptide synthesized. The work is performed accordingly to the Russian Government Program of Competitive Growth of Kazan Federal University; by the subsidy allocated to Kazan Federal University for the project part of the state assignment in the sphere of scientific activities and also supported by Russian Foundation for Basic Research (Grant 14-04-31029 mol_a).
Author information
Authors and Affiliations
Corresponding author
Additional information
Database Structural data are available in the Protein Data Bank/BioMagResBank databases under the accession numbers 2MZ6/25474.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Usachev, K.S., Efimov, S.V., Kolosova, O.A. et al. Antimicrobial peptide protegrin-3 adopt an antiparallel dimer in the presence of DPC micelles: a high-resolution NMR study. J Biomol NMR 62, 71–79 (2015). https://doi.org/10.1007/s10858-015-9920-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-9920-0