Abstract
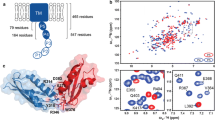
The insertase BamA is the central protein of the Bam complex responsible for outer membrane protein biogenesis in Gram-negative bacteria. BamA features a 16-stranded transmembrane β-barrel and five periplasmic POTRA domains, with a total molecular weight of 88 kDa. Whereas the structure of BamA has recently been determined by X-ray crystallography, its functional mechanism is not well understood. This mechanism comprises the insertion of substrates from a dynamic, chaperone-bound state into the bacterial outer membrane, and NMR spectroscopy is thus a method of choice for its elucidation. Here, we report solution NMR studies of different BamA constructs in three different membrane mimetic systems: LDAO micelles, DMPC:DiC7PC bicelles and MSP1D1:DMPC nanodiscs. The impact of biochemical parameters on the spectral quality was investigated, including the total protein concentration and the detergent:protein ratio. The barrel of BamA is folded in micelles, bicelles and nanodiscs, but the N-terminal POTRA5 domain is flexibly unfolded in the absence of POTRA4. Measurements of backbone dynamics show that the variable insertion region of BamA, located in the extracellular lid loop L6, features high local flexibility. Our work establishes biochemical preparation schemes for BamA, which will serve as a platform for structural and functional studies of BamA and its role within the Bam complex by solution NMR spectroscopy.






Similar content being viewed by others
Abbreviations
- DMPC:
-
1,2-Dimyristoyl-sn-glycero-3-phosphocholine
- DiC7PC:
-
1,2-Diheptanoyl-sn-glycero-3-phosphocholine
- LDAO:
-
Lauryldimethylamine-N-oxide
- POTRA:
-
Polypeptide transport-associated
- BAM:
-
β-barrel assembly machinery
- OMP:
-
Outer membrane protein
References
Albrecht R, Zeth K (2011) Structural basis of outer membrane protein biogenesis in bacteria. J Biol Chem 286:27792–27803
Albrecht R, Schütz M, Oberhettinger P, Faulstich M, Bermejo I, Rudel T, Diederichs K, Zeth K (2014) Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D 70:1779–1789
Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J Biomol NMR 6:1–10
Bayburt TH, Grinkova YV, Sligar SG (2002) Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Lett 2:853–856
Bos MP, Robert V, Tommassen J (2007) Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep 8:1149–1154
Browning DF, Matthews SA, Rossiter AE, Sevastsyanovich YR, Jeeves M, Mason JL, Wells TJ, Wardius CA, Knowles TJ, Cunningham AF, Bavro VN, Overduin M, Henderson IR (2013) Mutational and topological analysis of the Escherichia coli BamA protein. PLoS One 8:e84512
Denisov IG, Grinkova YV, Lazarides AA, Sligar SG (2004) Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J Am Chem Soc 126:3477–3487
Dong C, Hou HF, Yang X, Shen YQ, Dong YH (2012a) Structure of Escherichia coli BamD and its functional implications in outer membrane protein assembly. Acta Crystallogr D 68:95–101
Dong C, Yang X, Hou HF, Shen YQ, Dong YH (2012b) Structure of Escherichia coli BamB and its interaction with POTRA domains of BamA. Acta Crystallogr D 68:1134–1139
Fairman JW, Noinaj N, Buchanan SK (2011) The structural biology of β-barrel membrane proteins: a summary of recent reports. Curr Opin Struct Biol 21:523–531
Gatzeva-Topalova PZ, Walton TA, Sousa MC (2008) Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16:1873–1881
Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC (2010) Structure and flexibility of the complete periplasmic domain of BamA: the protein insertion machine of the outer membrane. Structure 18:1492–1501
Green MR, Sambrook JF (2012) Molecular cloning: a laboratory manual, 4th edn. Cold Spring Harbor Laboratory Press, New York
Gruss F, Zähringer F, Jakob RP, Burmann BM, Hiller S, Maier T (2013) The structural basis of autotransporter translocation by TamA. Nat Struct Mol Biol 20:1318–1320
Güntert P, Dötsch V, Wider G, Wüthrich K (1992) Processing of multi-dimensional NMR data with the new software PROSA. J Biomol NMR 2:619–629
Hagan CL, Kim S, Kahne D (2010) Reconstitution of outer membrane protein assembly from purified components. Science 328:890–892
Höhr AI, Straub SP, Warscheid B, Becker T, Wiedemann N (2015) Assembly of β-barrel proteins in the mitochondrial outer membrane. Biochim Biophys Acta 1853:74–88
Kay LE, Torchia DA, Bax A (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28:8972–8979
Kelly AE, Ou HD, Withers R, Dötsch V (2002) Low-conductivity buffers for high-sensitivity NMR measurements. J Am Chem Soc 124:12013–12019
Kim KH, Paetzel M (2011) Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J Mol Biol 406:667–678
Kim S, Malinverni JC, Sliz P, Silhavy TJ, Harrison SC, Kahne D (2007) Structure and function of an essential component of the outer membrane protein assembly machine. Science 317:961–964
Kim KH, Aulakh S, Tan W, Paetzel M (2011a) Crystallographic analysis of the C-terminal domain of the Escherichia coli lipoprotein BamC. Acta Crystallogr F 67:1350–1358
Kim KH, Kang HS, Okon M, Escobar-Cabrera E, McIntosh LP, Paetzel M (2011b) Structural characterization of Escherichia coli BamE, a lipoprotein component of the β-barrel assembly machinery complex. Biochemistry 50:1081–1090
Kim KH, Aulakh S, Paetzel M (2012) The bacterial outer membrane β-barrel assembly machinery. Protein Sci 21:751–768
Kjaergaard M, Poulsen FM (2011) Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution. J Biomol NMR 50:157–165
Lee D, Hilty C, Wider G, Wüthrich K (2006) Effective rotational correlation times of proteins from NMR relaxation interference. J Magn Reson 178:72–76
Leonard-Rivera M, Misra R (2012) Conserved residues of the putative L6 loop of Escherichia coli BamA play a critical role in the assembly of β-barrel outer membrane proteins, including that of BamA itself. J Bacteriol 194:4662–4668
Malinverni JC, Werner J, Kim S, Sklar JG, Kahne D, Misra R, Silhavy TJ (2006) YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol 61:151–164
McMorran LM, Brockwell DJ, Radford SE (2014) Mechanistic studies of the biogenesis and folding of outer membrane proteins in vitro and in vivo: What have we learned to date? Arch Biochem Biophys (In press) doi:10.1016/j.abb.2014.1002.1011
Ni D, Wang Y, Yang X, Zhou H, Hou X, Cao B, Lu Z, Zhao X, Yang K, Huang Y (2014) Structural and functional analysis of the β-barrel domain of BamA from Escherichia coli. FASEB J 28:2677–2685
Noinaj N, Kuszak AJ, Gumbart JC, Lukacik P, Chang H, Easley NC, Lithgow T, Buchanan SK (2013) Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501:385–390
Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK (2014) Lateral opening and exit pore formation are required for BamA function. Structure 22:1055–1062
Onufryk C, Crouch ML, Fang FC, Gross CA (2005) Characterization of six lipoproteins in the σE regulon. J Bacteriol 187:4552–4561
Pervushin K, Riek R, Wider G, Wüthrich K (1997) Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci USA 94:12366–12371
Raschle T, Hiller S, Yu TY, Rice AJ, Walz T, Wagner G (2009) Structural and functional characterization of the integral membrane protein VDAC-1 in lipid bilayer nanodiscs. J Am Chem Soc 131:17777–17779
Rigel NW, Silhavy TJ (2012) Making a β-barrel: assembly of outer membrane proteins in gram-negative bacteria. Curr Opin Microbiol 15:189–193
Salzmann M, Pervushin K, Wider G, Senn H, Wüthrich K (1998) TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA 95:13585–13590
Selkrig J, Leyton DL, Webb CT, Lithgow T (2014) Assembly of β-barrel proteins into bacterial outer membranes. Biochim Biophys Acta 1843:1542–1550
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol Syst Biol 7:539
Sinnige T, Weingarth M, Renault M, Baker L, Tommassen J, Baldus M (2014) Solid-state NMR studies of full-length BamA in lipid bilayers suggest limited overall POTRA mobility. J Mol Biol 426:2009–2021
Sklar JG, Wu T, Gronenberg LS, Malinverni JC, Kahne D, Silhavy TJ (2007) Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA 104:6400–6405
Voulhoux R, Tommassen J (2004) Omp85, an evolutionarily conserved bacterial protein involved in outer-membrane-protein assembly. Res Microbiol 155:129–135
Walther DM, Rapaport D, Tommassen J (2009) Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes: evolutionary conservation and divergence. Cell Mol Life Sci 66:2789–2804
Warner LR, Varga K, Lange OF, Baker SL, Baker D, Sousa MC, Pardi A (2011) Structure of the BamC two-domain protein obtained by Rosetta with a limited NMR data set. J Mol Biol 411:83–95
White SH (2004) The progress of membrane protein structure determination. Protein Sci 13:1948–1949
Wider G, Dreier L (2006) Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc 128:2571–2576
Yu TY, Raschle T, Hiller S, Wagner G (2012) Solution NMR spectroscopic characterization of human VDAC-2 in detergent micelles and lipid bilayer nanodiscs. Biochim Biophys Acta 1818:1562–1569
Zhang H, Gao ZQ, Hou HF, Xu JH, Li LF, Su XD, Dong YH (2011) High-resolution structure of a new crystal form of BamA POTRA4–5 from Escherichia coli. Acta Crystallogr F 67:734–738
Zhou P, Lugovskoy AA, Wagner G (2001) A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR 20:11–14
Zhu G, Xia Y, Nicholson LK, Sze KH (2000) Protein dynamics measurements by TROSY-based NMR experiments. J Magn Reson 143:423–426
Acknowledgments
We thank Sina Reckel and Thomas Raschle for technical advice and discussions. This work was supported by grants from the Swiss National Science Foundation (Grant PP00P3_128419) and the European Research Council (FP7 contract MOMP 281764) to S.H. The sequence-specific resonance assignments of BamA in lipid bilayer nanodiscs (residues D678–Q698) were deposited in the BMRB (accession code 25359).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Morgado, L., Zeth, K., Burmann, B.M. et al. Characterization of the insertase BamA in three different membrane mimetics by solution NMR spectroscopy. J Biomol NMR 61, 333–345 (2015). https://doi.org/10.1007/s10858-015-9906-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10858-015-9906-y