Abstract
Despite large numbers of reintroduction projects taking place and the high cost involved, there is a generally low success rate. Insects in particular are understudied within reintroduction ecology, with guidelines focusing on more iconic vertebrate taxa. Species distribution models (SDMs) examine the associations between species observations and environmental variables to find the conditions in which populations could survive. This study utilises two frequently used SDM approaches, a regression model (general linear model (GLM)) and a machine learning method (MaxEnt) to model habitat suitability for Chequered Skipper, Carterocephalus palaemon, butterflies, which are being reintroduced to Northamptonshire following extinction in England. We look at how SDMs using widespread remotely sensed variables could be used to inform the reintroduction process by finding areas of suitable habitat that were previously overlooked. These remotely sensed variables have the potential to inform reintroductions without extensive on the ground research as they cover huge areas and are widely available. We found that both models are successful in discriminating between presences and absences, using only a limited number of explanatory variables. We conclude that these wide-scale SDMs are useful as a first step in the decision-making process in determining appropriate sites for reintroductions, but that they are less accurate when establishing precisely where species should be placed.
Implications for insect conservation
The rates of failure among species reintroductions are very high despite the large costs involved in these projects. By better utilising SDMs and remote sensing variables that cover huge areas, an increased rate of success and cost efficiency in insect reintroductions could be established.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Habitat loss and fragmentation caused by land use changes is one of the leading challenges many species are facing. Species that are unable to move long distances are particularly susceptible to suffering from habitat loss and fragmentation of meta-populations (Thomas et al. 2001). When distances between populations or suitable habitats become greater than the dispersal capacity of the species, genetic exchange and natural colonisation (or recolonisation) are unlikely or often impossible (Maes et al. 2019). Reintroductions, defined as ‘the intentional movement and release of an organism inside its indigenous range from which it has disappeared’ (IUCN/SSC 2013), of individuals from sufficiently large populations to suitable areas can be applied as a conservation measure to help tackle the effects of habitat fragmentation and loss (Byrne and Pitchford 2016). The need for reintroductions as a conservation measure has therefore never been higher, with many species potentially being able to benefit. However, reintroductions have historically had a generally low success rate (Bubac et al. 2019). Despite large numbers of reintroduction projects taking place, the science that underpins reintroduction biology is still being developed (Armstrong and Seddon 2008; Berger-Tal et al. 2020; Taylor et al. 2017). Successful cases of insect reintroductions, such as the case of the Large Blue Butterfly (Phengaris arion Linnaeus, 1758) provide evidence of the potential of reintroduction programs in insects (Andersen et al. 2014; Ellis et al. 2011; Thomas et al. 2009). However, most of the published work is restricted to a case-by-case basis and largely specific to one species (Oates and Warren 1990), leading to a lack of a theoretical framework in which reintroductions can be based (Byrne and Pitchford 2016). This issue is particularly prevalent among insect species, where there are much fewer studies on reintroductions, making it difficult to establish a framework for insects specifically.
In order to ensure that an area is suitable for reintroductions it is important to understand the relationships between species and their environment (Osborne and Seddon 2012). Species distribution models (SDMs), also referred to as habitat suitability models, examine the associations between species observations and environmental variables to find the conditions in which populations could potentially survive (Elith and Leathwick 2009). SDMs are often used to locate species occurrences or to identify areas that could be important for conservation action (Villero et al. 2017). With the increasing impact of climate change, SDMs have been gaining in importance, with several studies using the models to predict climate-driven changes in species ranges (Bond et al. 2011; Fourcade 2016). SDMs are also widely used in the context of species reintroductions (Bellis et al. 2020; Hunter-Ayad et al. 2020; Smeraldo et al. 2017), supporting decision makers in regard to land use planning and in determining suitable locations for habitat restoration and the reintroductions themselves. However, there is some question over whether SDMs can accurately predict suitable habitat in areas beyond the area used to train the model, limiting their use in species reintroductions to smaller areas. This issue could be overcome with the improvement of wide scale remote sensing that allow environmental variables to be taken from vast ranges, even worldwide, in a comparable manner. Some currently widely used remote sensing products have several limitations that reduce their ability to be used successfully, such as either low spatial detail, and an under-representation of smaller land patches or not covering wide enough areas for use in species that range across several countries (Rosina et al. 2018). However, remotely sensed data that is easily accessible, has high spatial resolution and is measured in a comparative way across large areas, is becoming much more common, potentially leading to huge improvements in the accuracy of the SDMs that use these types of data. The use of SDMs in choosing sites is often supplemented with knowledge from species experts in several ways, particularly in making the final decisions on sites (Maes et al. 2019). This becomes an issue in species that have not been extensively studied or where there are very few experts, a particular problem amongst species considered less charismatic, including many insects (Hochkirch et al. 2023). The use of wide-scale remote sensing data, that does not necessarily require extensive previous knowledge of the species’ environmental requirements, could open up the number of species that could benefit from reintroductions.
There is currently a diverse range of analytical approaches used by ecologists to carry out species distribution models. Two of the most frequently used are regression models and machine learning methods (Shabani et al. 2016). Regression models such as generalised linear models (GLM) use the relationship between environmental variables and presence and absence points. Machine learning methods create predictions based upon rules created by these relationships between observations and environmental conditions (Vollering et al., 2019). An example is MaxEnt, which compares probability densities from presence locations and background points to derive the probability of occurrence (Merow et al. 2013). This study utilises GLMs and MaxEnt to model predicted habitat suitability of Chequered Skipper (Carterocephalus palaemon Pallas, 1771) butterflies. The most significant difference in the use of these two models is that the GLM can use both presence/absence data whereas the MaxEnt uses presence-only. Presence/absence data can be more difficult to obtain and is not available for all species but is thought of as the preferred method and has been found to be accurate in predicting species distributions (Brotons et al. 2004). By comparing these two methods using a larger, more easily obtained dataset of presence points for the MaxEnt and a smaller dataset of absence/presence points for the GLM, we can assess whether the addition of absence points does make a noticeable difference in the accuracy of the models.
As part of the ‘Back from the Brink’ programme, the charity Butterfly Conservation began a reintroduction project for Chequered Skipper to parts of England, taking individuals from the Wallonia (southern) region of Belgium and translocating them into England. Across Europe, Chequered Skippers reside in a variety of habitats including heathland and grasslands, although usually found at woodland edges in wide rides and glades (Maes et al. 2019). Chequered Skippers use grass hostplants across Europe including purple moor-grass, Molinia caerulea, false brome, Brachypodium sylvaticum, heath false brome, Brachypodium pinnatum and Yorkshire fog, Holcus lanatus (Moore 2004; Weidemann 1988). In historical English populations, both Brachypodium spp were thought to be the main hostplants (Emmet and Heath 1989). The Chequered Skipper was always considered rare in Britain, in England it was confined to a band of woods and associated limestone grasslands in the East Midlands, from Oxford to Lincolnshire but became extinct here in 1976, likely through changes in habitat management, land use changes and encroachment of its habitat (Moore 2004). Within Britain, the species is now confined to a small area of western Scotland (Butterfly Conservation 2019). The project aimed to reintroduce the Chequered Skipper to several connected sites within an area to establish a functioning meta-population (Maes et al. 2019). This is in line with the results of previous projects that have applied metapopulation theory to landscape scale planning of butterfly conservation and found that reintroduction to multiple sites is more likely to succeed (Ellis et al. 2011). Here, we use Chequered Skipper as an exemplar insect species, to explore what might happen if reintroductions need to be based on relatively generic data. We do not aim to mimic or rely on the decision-making that led to the current re-introduction into one of the species’ historical strongholds, and where the selection of source populations was based on a set of landcover-based SDMs (Maes et al. 2019).
The aim of this study is to look at the role that SDMs that use wide scale remotely sensed data could play in informing the reintroduction process by finding areas of suitable habitat that Chequered Skippers could move into as they spread through the landscape, locating areas of habitat that were previously unknown to practitioners. On a broader scale, it will also look at whether the use of these widely sensed habitat variables in SDMs could reduce the requirement of extensive species specific knowledge in choosing sites, therefore expanding the number of species that could benefit from reintroductions without the need for years of extensive studying.
The key questions asked are:
-
1.
Using two different modelling approaches, one of which uses absence and presence data and one of which uses presence-only data (GLM and MaxEnt respectively), and widespread remotely sensed climate and environmental variables, is it possible to predict the recent distribution of Chequered Skippers?
-
2.
How can the use of species distribution models improve success in choosing areas for reintroductions and supplement expert knowledge, particularly for lesser-known taxa?
-
3.
How do the advantage and disadvantages of these model types and their outputs affect their use in reintroduction projects?
Methods
Study species and data
Data on the presence of Chequered Skipper was taken from two sources: the European Butterfly Monitoring Scheme, eBMS, (European Butterfly Monitoring Scheme 2020) for the GLM analysis and the Global Biodiversity Information Facility, GBIF (GBIF.org 2019),for the MaxEnt analysis. eBMS is a standardised, long running, transect based survey on all European butterfly species carried out mostly by volunteers across most of Europe. The records used in this analysis covered four countries: Belgium, France, Germany and the Netherlands. All data is first collected at a national level and validated by eBMS to ensure accuracy. Validation methods include investigating submissions that fall outside a species known range or flight period and using photographic evidence from volunteers. Transects completed from 1990 to 2019 were included in the data set. Duplicate transects at single sites were removed to create presence/absence only at each of the 4274 individual sites, 493 of which had Chequered Skippers present. Chequered Skippers were indicated as present at a transect location if they had been seen at the location within the time period. Absence was inferred when transects had been walked at that location, but no Chequered Skippers were seen.
GBIF is an international network and data infrastructure aiming to provide open access to data from a network of sources across the world that use common standards and open-source tools to share information about where and when species have been recorded (Telenius 2011). GBIF does not provide transect information so provides presence data only. All presence data (11,900 observations) from 1990 to 2019 was included in the analysis and covered the same four countries: Belgium, France, Germany and the Netherlands.
Landscape scale habitat factors and climate factors likely to be influential were first identified through previous knowledge of the species’ ecology (Maes et al. 2019; Moore 2004). We started with nine possible predictor variables for both models: tree cover (average cover over 100 m), tree cover (20 m), tree cover2 (100 m) to allow for a non-linear correlation with tree cover, leaf area index, precipitation in the driest quarter, soil moisture (Copernicus, 2019) and average annual temperature, temperature in the hottest quarter (Fick and Hijmans 2017). For the GLM, environmental data was extracted from raster layers and bound to eBMS butterfly presence/absence data using the packages raster (Hijmans 2022) and rgdal (Bivand et al. 2021). For the MaxEnt model the raster layers of environmental variables were transformed using ArcGIS to be the same resolution of 1km2 and extent and then stacked into a single raster stack, with the GBIF data entered as points. Higher resolution variables were averaged per square to fit the variable of the lowest resolution. As a result, only one version of tree-cover (100 m) was tested in this model.
Model calibration (GLM)
We developed a generalised linear model in R (version 3.6.2) to assess the relationship between environmental variables and species presences/ absences obtained from eBMS data from the aforementioned four countries, as this dataset contained true absences. We used the package dismo (Hijmans et al. 2020) to develop the model. The model was set as binomial as the butterfly data is binary (either present or absent). Packages rgdal (Bivand et al. 2021) and maptools (Bivand and Lewin-Koh 2021) were used to create habitat suitability maps.
In order to choose the most appropriate set of environmental predictors, we used a backwards stepwise approach considering model performance (assessed through AUC on a test set of data of presences and absences), starting out with the 9 predictor variables. 20% of the data was removed randomly before model building to be used as a validation set. Model performance was also used to establish the importance of each variable in the final model based on the change in AUC between the variables removal and inclusion in the model.
Model calibration (MaxEnt)
To run our model we used MaxEnt within R which relies on the following R packages: dismo (Hijmans et al. 2020), raster (Hijmans 2022), rgeos (Bivand and Rundel 2021) and rJava (Urbanek 2021). The MaxEnt algorithm compares presence locations and variable interactions to similar interactions of background locations, which are randomly selected cells (by default 10,000) in which presence is not known and does not have missing data in any of the predictor variables (Feng et al. 2017). We started with the same set of nine environmental variables as the GLM and we used the same stepwise approach considering AUC (based on a validation set of data randomly removed before model building and using 1000 randomly chosen points as pseudo-absences). MaxEnt models use their own stepwise process to determine the percentage contribution of each variable within the model which was used for assessment of variable importance in the final model (Merow et al. 2013).
Threshold metrics
Thresholds were used to convert continuous predictions into binary predictions of areas of habitat and non-habitat. We used the average probability approach to determine threshold values for each model, taking the average predicted occurrence probability of the model-building data (specifically the presence data) as the threshold (following recommendations in Liu et al. 2005).
Model prediction evaluation and comparisons
To evaluate how well the models predict suitable habitat we compared the outputs to known distributions in the Wallonia region of Belgium. We compared them to a distribution of 1569 presence points in this region (provided by Public Service of Wallonia), using the threshold metrics to determine whether the presence points fell into suitable habitat or non-habitat locations according to the model. We used this area to test the model as it did not contain any eBMS or GBIF data points and independent of data used in the model building.
We then additionally compared both models’ predictions to the eBMS dataset across all four countries, looking at the average and range of predicted suitability at known presence and absence points. We also compared the model predictions on this dataset using AUC to compare how well the models distinguished between presences and absences. Finally, to facilitate our discussion of re-introduction possibilities, we plotted model predictions to England and visually compared them to the pre-extinction (1856–1976) English distribution as per the Butterflies for the New Millennium (BNM) project dataset (Butterfly Conservation 2019). This included highlighting areas that are predicted to be suitable habitat by both models (effectively a consensus model ensemble (Araújo and New 2007), indicating that there is less uncertainty in the suitability of these areas.
Results
Predictor variables
The GLM variable selection procedure identified four predictors (Table 1) as those producing the best models. Five variables were selected for use in the final MaxEnt model (Table 1).
The importance of each variable within the two models was also determined to be slightly different. The order of importance from the GLM was determined through a stepwise process of removing and adding individual variables to the model and the reduction in the AUC value when the variable is removed from the model. The order of importance as determined by the GLM was: leaf area index, tree-cover, precipitation in the driest quarter and soil moisture. All variables in the GLM had a positive relationship with habitat suitability and so have higher values in areas above the threshold values for suitable habitat than non-suitable habitat.
MaxEnt models use their own stepwise process to determine the percentage contribution of each variable within the model through the removal and then inclusion of individual variables. The relative importance of each variable according to the MaxEnt model in order of importance according to percentage contribution was: annual mean temperature, tree-cover, soil moisture, max temperature and precipitation in the driest quarter. Response curves indicated a positive relationship with habitat suitability for tree-cover, soil moisture and precipitation. Max temperature (Bio5) had a negative relationship with habitat suitability, steeply declining above approximately 37 °C. Mean temperature (Bio1) had a positive relationship with habitat suitability to around 30 °C and then steeply declined.
Habitat suitability maps
Both the GLM and MaxEnt models show similar spatial patterns of predicted habitat suitability, although the MaxEnt model shows a much higher habitat suitability in the majority of locations. Habitat suitability as predicted by the GLM and MaxEnt model is shown in Fig. 1, at three different spatial scales in Belgium. We have focused these results on Belgium, as this includes the chosen source region for the reintroduction. High predicted suitability is shown across a strip in the Wallonia region, which contains the locations of the populations used for the English reintroduction. Suitability (as established by different thresholds for each model determined by the average prediction value of the model building presence data) was compared to a known location of individuals across Wallonia, from a dataset not used to train the models (Fig. 2). 78% of the presence locations in Wallonia were in the high suitability bracket as predicted by the GLM (defined by the threshold), whereas only 58% were in the high suitability bracket as predicted by the MaxEnt model.
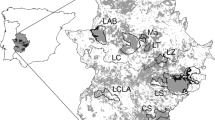
Using Europe-wide consistent variables has allowed us to make predictions of suitability for Britain. Figure 3 shows the predicted potential distribution in the east midlands of England. South and central Britain tends to have a slightly lower range of suitability values than Belgium. Values in the high suitability bracket cover 5.4% of the land according to the GLM or 11.1% according to MaxEnt (Fig. 4). 4.5% of the land is highly suitable according to both models. The last stronghold for the species in England before its extinction, as shown in Fig. 4 which shows model predictions compared to known historical presences in east-central England, is assigned as patchily suitable habitat by both models. Most of the suitable habitat in these areas is highlighted by the MaxEnt model, with only a small amount being deemed suitable by both, and comprise small individual sites rather than large areas of connected suitable habitat. The areas of highest predicted suitability in south-east England contain only a couple of historical records of Chequered Skipper.
Habitat suitability maps of Chequered Skippers in Belgium. These are shown at three scales going from top to bottom: Belgium wide, Wallonia (the source area) and zoomed in closer to a small area that contains some of the source populations. a-c show results for the GLM and d-f the results of the MaxEnt model, both shown at the same three scales of the same area. The maps use the same colour ramp for both modelling methods on a scale of 0–1, with 1 being the highest possible suitability
Predicted areas of habitat in England taken from threshold values for each model. Areas highlighted as habitat by each model are shown as orange (MaxEnt) and blue (GLM). Areas that would be deemed most suitable habitat, as they are highlighted as habitat by both models, are shown in red. The known historical distribution (including records dating from 1854–1976) is shown as black dots
Model comparison
Figure 5 shows the predictions of both models at eBMS presence and absence points to show how well they predict a known distribution. Both models are successful at discriminating presences from absences (AUC 0.801 and 0.76, GLM and MaxEnt respectively when compared to the whole eBMS dataset). However, the MaxEnt model predicts higher values for both than the GLM. The GLM gives a mean predicted habitat suitability at known presence points at 0.261 compared to an average of 0.098 at absence points. Whereas the MaxEnt model gave a mean suitability at presence points of 0.747 compared to 0.505 at absence points. Despite this, as can be seen in Figs. 2 and 3(a and b), both models pick out the same general locations as having more suitable habitats. This was confirmed using a Spearman’s rho correlation coefficient on the relationship between the predictions from both models taken at the same points across Europe which resulted in a significant positive correlation (rs=0.72, p < 0.001).
Discussion
GLM vs. MaxEnt: comparisons of method outputs and advantages
Overall, both models suggest similar areas as having the highest suitability for Chequered Skipper. In this study we computed AUC of both models using the dataset of presence and absences, giving an AUC of 0.801 and 0.78 for GLM and MaxEnt respectively. The MaxEnt model predicts higher values for both absence and presence points than the GLM, however their spatial patterns are concordant and the numerical predictions highly correlated (rs=0.72).
This difference in ‘scale’ of the predicted suitability is likely due to the GLM containing absences and MaxEnt being a presence-only algorithm, using ‘pseudo-absences’ or background points rather than true absences. This is precisely what is thought to be one of the main drawbacks of MaxEnt (Yackulic et al. 2013). Using background points rather than true absences is also thought to have a number of disadvantages, including difficulties in model calibration (especially using AUC) and less clear threshold selection when converting model output into suitable/unsuitable locations (as there is no way to balance false-positive and false-negative predictions) (Brotons et al. 2004). In general, interpretation of the meaning of background data or pseudoabsence data varies throughout literature, making it difficult to interpret their outputs meaningfully (Elith and Leathwick 2009).
The differences in presence-only and presence absence models was also evident in the selection of model variables. While both models used tree-cover (100 m), soil moisture and precipitation, both temperature variables (mean temperature and maximum temperature) were selected for use in the MaxEnt model but not in the GLM, whereas leaf area index was only selected in the GLM. Temperature variables may not have been selected for in the GLM model, as the areas in which presence and absence points were located across Europe did not differ greatly in temperature: both presence and absence points were located in similar regions, or in some cases the same regions of area. However, this may not have been picked up in the presence only models, where it only takes into account the fact that all presence points were located in areas with similar temperatures.
Despite the negatives of MaxEnt that come from it being presence-only, it is still useful in picking out locations of suitable habitat, and this is supported by the fact that it picks out similar areas to our presence/absence GLM. MaxEnt is very popular due to its easy applicability and ability to be used in combination with R (Merow et al. 2013). The MaxEnt algorithm is particularly robust relating to irregular distributed small sample sizes and this makes it especially interesting for modelling rare species (Yackulic et al. 2013). Additionally, some argue that absence data can be misleading when the species or environment is not at equilibrium (e.g., invasions, climate change) or the species is not easy to find/study (Elith and Leathwick 2009), possibly introducing confounding information because they can indicate either habitat that is unsuitable or habitat that is suitable but currently unoccupied (Elith and Leathwick 2009; Jiménez-Valverde et al. 2008).
Setting thresholds for suitable habitat is one way to easily compare maps of habitat suitability between the two methods. We tested the effectiveness of predicting habitat suitability in areas where there were no training data by comparing predicted suitability to a known distribution in the Wallonia region of Belgium. Using threshold values determined by the values given at points from the training data we found that the GLM predicted that the habitat would be suitable at 78% of the locations but only 58% of the locations were predicted to be in suitable habitat as shown by the MaxEnt model. This suggests that the GLM is better at predicting areas of suitable habitat, but it is worth noting that this is only compared to presence points and so it can only give an indication of accuracy rather than a definitive answer. Interestingly, the MaxEnt predicts that a larger percentage of south-eastern England contains suitable habitat than the GLM. This difference could come from the difference in how important each variable is considered within the separate models, leading them to pick out different areas as suitable habitat.
In general, when good quality presence/absence data is available, using a regression method such as GLMs is the preferred method and has been found to be accurate in predicting species distributions (Brotons et al. 2004). In our case, despite using much fewer points for the training data, the GLM more accurately predicts presence locations in Belgium, correctly predicting the areas as habitat in 78% of the sites compared to MaxEnts 63%. However, it is worth noting that good quality presence/absence data is often not available. Machine learning methods such as MaxEnt that use presence and pseudoabsences are not far behind in their ability to distinguish suitable habitat. Additionally, in this instance it is only a comparison to presence locations, which can only tell some of the story. Our results, particularly the high correlation between GLM and MaxEnt predictions, suggest that the use of the species’ presence-only distribution modelling such as MaxEnt is still useful for defining the suitable habitat (Yackulic et al. 2013). In cases where information on absences of the species is lacking, these methods are certainly still a good replacement for presence-absence models. If looking for a method to use on an understudied species, in reality a presence-only model is likely to be the only method available for use (Warren et al. 2020; Yackulic et al. 2013).
The use of SDMs in species reintroductions
Both models are successful in discriminating between presences and absences, using only four/five explanatory variables. This indicates that both models could be potentially useful in choosing sites for reintroduction that could provide the suitable habitat required. One of the best components of both models is that they use remote sensing data that covers extremely large areas, with each of the predictor variables crossing either all of Europe or even the world and being quantified with the same method across the whole area (Copernicus, 2019). One of the difficulties in using SDMs in reintroduction is that it is thought risky to predict habitat suitability outside the physical range in which the model was trained (Velazco et al. 2023), creating an issue in cases where species are being reintroduced to old ranges far outside of their current range (Jarvie and Svenning 2018). By using predictor variables that are comparable across widespread areas and several countries, this potentially removes one of the issues of SDMs for use in reintroductions.
On the other hand, being limited by the availability of wide-extent variables could potentially lose some of the power of a model. Some important variables could be missed, in particular in regard to details of the ecological community. Variables such as host plants used could differ across countries. In this case, it is known that Chequered Skippers use a variety of hostplants across Europe, with potentially different populations using different specific hostplants. Furthermore, as well as not being detectable with remote sensing, most ecological community variables aren’t surveyed consistently across countries. For example, while it is clear that the models do predict the current distribution well in places such as the Wallonia region of Belgium, they are not perfect in their predictions and in places predict habitat outside the regions where we know Chequered Skippers are found. This is likely because of the limits in the variables that can be incorporated in such wide scale studies. Whilst additional macroclimate variables did not improve our models (see methods), it is likely that small scale edaphic, microclimate and habitat composition factors, such as hostplants, influence the ability of Chequered Skipper to survive within the broad areas identified as suitable according to the four variables in our models.
Additional to choosing areas, these models can help define what suitable habitat for the species is, and potentially help predict how the species may react to environmental changes or events in the years after the reintroduction has taken place (Bellis et al. 2023). For example, tree-cover, averaged over 100 m, was a critical variable within both models. Tree-cover at a scale of 20 m however, was not significant and removed from the models, possibly accounting for the fact that the species resides on glades and rides within forests and on forest edges, and therefore thick tree-cover in the immediate area is not an important factor, whereas tree-cover in the wider area is. Precipitation and soil moisture were important variables in both models and both had a positive relationship with habitat suitability. As precipitation is taken from the driest three months of the year and we chose to use soil moisture values from May-June 2018 (a period of time where many European countries were experiencing drought), this indicates that Chequered Skippers are susceptible to drought. While much of this specific habitat information was known previous to the introduction of Chequered Skippers into England, the results show the use of SDMs is pinpointing specific wide-scale habitat requirements for species in reintroductions and in predicting how a species may be affected by extreme climate events (Bellis et al. 2023). This would be particularly useful in understudied species, which is the case for many insects, where extensive studying of habitat requirements has not taken place.
The models suggest that south-east England and eastern England contain some highly suitable areas, which were unexpected as suggestions for re-introduction areas as previous records do not indicate that the species occurred there. This suggests that it may not have provided the suitable habitat required for the species, putting into question the performance of the models in these areas. Furthermore, in the region that historical records show to be the previous stronghold in England (Asher et al. 2001), the majority of predicted suitable habitat is from the MaxEnt model only, with very few places predicted to be suitable by the GLM alone or both models. These locations are also very patchy, looking to be single sites of suitable habitat instead of large areas of interconnected sites as shown further south. This could again indicate a lack of accuracy in the models, particularly the GLM, for regions outside the source data, and indicates that some caution should be taken in the use of these models for reintroductions that are far outside the source data regions (Velazco et al. 2023). However, the fact that there is little historical evidence that the species ever occurred there doesn’t mean they are not biophysically suitable currently. The species was likely under-recorded historically because there are widely scattered singleton records. Additionally, small patchy sites that lack connectivity could have been part of what led to the extinction of the Chequered Skipper in the first place. In this context, the predictions from both models of only small and patchy areas of suitable habitat are somewhat consistent with the extinction of the species in England. Practitioners planning an expensive re-introduction programme would likely seek a broader range of evidence sources before committing to any particular area. Our results could be a useful prompt to seek that evidence, and look for opportunities to maximise the range of the species in future rather than being limited to re-establishing a known historical range.
Other studies have found that the most successful methods in translocations combine both SDMs and in situ field validation (Draper et al. 2019). This suggests that while SDMs such as these are useful in picking out suitable habitat to some extent, it does not remove the need for experts on the species and therefore for the species to be highly studied before the reintroduction, such as with the Chequered Skipper (Maes et al. 2019). SDMs in species reintroductions definitely have their place in determining appropriate sites, but cannot be relied upon completely to establish where species should be placed, potentially missing the finer scale detail in finding suitable sites. In the case of the Chequered Skipper, this study has shown that they could have use in pinpointing potential areas for the species to expand into. However, this will need to be backed up with on the ground knowledge and potentially field validation of new sites.
Data availability
The presence/absence data is accessible in European Butterfly Monitoring Scheme (eBMS) https://butterfly-monitoring.net/ with permission from individual national butterfly monitoring schemes. Presence-only data is freely available from Global Biodiversity Information Facility (GBIF) https://www.gbif.org/. All environmental data is freely available from Copernicus Global Land Service https://land.copernicus.eu/global/products and Worldclim https://www.worldclim.org/data/index.html. Model validation datasets were provided by Butterfly conservation https://doi.org/10.15468/tqf8z3 and Public Service of Wallonia http://observatoire.biodiversite.wallonie.be/ through request. Habitat suitability maps at a European scale are available at https://github.com/ghalford6/Chequered_Skipper_habitat_maps.git.
References
Andersen A, Simcox DJ, Thomas JA, Nash DR (2014) Assessing reintroduction schemes by comparing genetic diversity of reintroduced and source populations: a case study of the globally threatened large blue butterfly (Maculinea arion). Biol Conserv 175:34–41. https://doi.org/10.1016/j.biocon.2014.04.009
Araújo MB, New M (2007) Ensemble forecasting of species distributions. Trends Ecol Evol 22:42–47. https://doi.org/10.1016/j.tree.2006.09.010
Armstrong DP, Seddon PJ (2008) Directions in reintroduction biology. Trends Ecol Evol 23:20–25. https://doi.org/10.1016/j.tree.2007.10.003
Asher J, Warren M, Fox R, Harding P, Jeffcoate G, Jeffcoate S (2001) The millennium atlas of butterflies in Britain and Ireland. Millenn. Atlas Butterflies Br. Irel
Bellis J, Bourke D, Maschinski J, Heineman K, Dalrymple S (2020) Climate suitability as a predictor of conservation translocation failure. Conserv Biol 34:1473–1481. https://doi.org/10.1111/cobi.13518
Bellis JM, Maschinski J, Bonnin N, Bielby J, Dalrymple SE (2023) Climate change threatens the future viability of translocated populations. Divers Distrib. https://doi.org/10.1111/ddi.13795
Berger-Tal O, Blumstein DT, Swaisgood RR (2020) Conservation translocations: a review of common difficulties and promising directions. Anim Conserv 23:121–131. https://doi.org/10.1111/ACV.12534
Bivand R, Lewin-Koh N (2021) maptools: Tools for Handling Spatial Objects. R package version 1.1-2
Bivand R, Rundel C (2021) rgeos: Interface to Geometry Engine - Open Source (‘GEOS’). R package version 0.5-8
Bivand R, Keitt T, Rowlingson B (2021) rgdal: Bindings for the Geospatial Data Abstraction Library. R package version 1.5–27
Bond N, Thomson J, Reich P, Stein J (2011) Using species distribution models to infer potential climate change-induced range shifts of freshwater fish in south-eastern Australia. Mar Freshw Res 62:1043. https://doi.org/10.1071/MF10286
Brotons L, Thuiller W, Araújo MB, Hirzel AH (2004) Presence-absence versus presence-only modelling methods for predicting bird habitat suitability. Ecography 27:437–448. https://doi.org/10.1111/j.0906-7590.2004.03764.x
Bubac CM, Johnson AC, Fox JA, Cullingham CI (2019) Conservation translocations and post-release monitoring: identifying trends in failures, biases, and challenges from around the world. Biol Conserv 238:108239. https://doi.org/10.1016/J.BIOCON.2019.108239
Butterfly Conservation (2019) Butterfly distributions for Scotland from Butterfly Conservation and the Biological records Centre. https://doi.org/10.15468/tqf8z3. accessed via GBIF.org Occurrence dataset
Byrne JGD, Pitchford JW (2016) Species reintroduction and community-level consequences in dynamically simulated ecosystems. Biosci Horiz Int J Stud Res 9. https://doi.org/10.1093/BIOHORIZONS/HZW009
Copernicus Land Monitoring Service (2019) European Environment Agency (EEA), European Union, accessed via https://www.copernicus.eu/en
Draper D, Marques I, Iriondo JM (2019) Species distribution models with field validation, a key approach for successful selection of receptor sites in conservation translocations. Glob Ecol Conserv 19:e00653. https://doi.org/10.1016/j.gecco.2019.e00653
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst 40:677–697. https://doi.org/10.1146/annurev.ecolsys.110308.120159
Ellis S, Wainwright D, Berney F, Bulman C, Bourn N (2011) Landscape-scale conservation in practice: lessons from northern England. UK J Insect Conserv 15:69–81. https://doi.org/10.1007/s10841-010-9324-0
Emmet AM, Heath J (eds) (1989) The moths and butterflies of Great Britain and Ireland, Volume 7, part 1: Hesperiidae—Nymphalidae (the butterflies). Harley, Colchester
European Butterfly Monitoring Scheme (2020) Accessed via: https://butterfly-monitoring.net/ebms
Feng X, Walker C, Gebresenbet F (2017) Shifting from closed-source graphical-interface to open-source programming environment: 1 a brief tutorial on running Maxent in R 2 3 introduction 26. https://doi.org/10.7287/peerj.preprints.3346v1
Fick SE, Hijmans RJ (2017) WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37:4302–4315. https://doi.org/10.1002/joc.5086
Fourcade Y (2016) Implication for predicting range shifts with climate change. Ecol Inf 36:8–14. https://doi.org/10.1016/j.ecoinf.2016.09.002. Comparing species distributions modelled from occurrence data and from expert-based range maps
GBIF.org (2019) Carterocephalus palaemon (Pallas, 1771) in GBIF Secretariat (2019). GBIF Backbone Taxonomy. Checklist dataset. https://doi.org/10.15468/39omei. accessed via GBIF.org
Hijmans R (2022) raster: Geographic Data Analysis and Modeling. R package version 3.5–15
Hijmans R, Phillips S, Leathwick J, Elith J (2020) dismo: Species Distribution Modeling. R package version 1.3-3
Hochkirch A et al (2023) A multi-taxon analysis of European red lists reveals major threats to biodiversity. PLoS ONE 18:e0293083. https://doi.org/10.1371/journal.pone.0293083
Hunter-Ayad J, Ohlemüller R, Recio MR, Seddon PJ (2020) Reintroduction modelling: a guide to choosing and combining models for species reintroductions. J Appl Ecol 57:1233–1243. https://doi.org/10.1111/1365-2664.13629
IUCN/SSC (2013) ‘Guidelines for reintroductions and other conservation translocations. Version 1.0’, Gland S IUCN Species Survival Commission, viiii + 57 p
Jarvie S, Svenning JC (2018) Using species distribution modelling to determine opportunities for trophic rewilding under future scenarios of climate change. Philos Trans R Soc B Biol Sci 373. https://doi.org/10.1098/rstb.2017.0446
Jiménez-Valverde A, Lobo JM, Hortal J (2008) Not as good as they seem: the importance of concepts in species distribution modelling. Divers Distrib 14:885–890. https://doi.org/10.1111/j.1472-4642.2008.00496.x
Liu C, Berry PM, Dawson TP, Pearson RG (2005) Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28:385–393. https://doi.org/10.1111/j.0906-7590.2005.03957.x
Maes D, Ellis S, Goffart P, Cruickshanks KL, van Swaay CAM, Cors R, Herremans M, Swinnen KRR, Wils C, Verhulst S, De Bruyn L, Matthysen E, O’Riordan S, Hoare DJ, Bourn NAD (2019) The potential of species distribution modelling for reintroduction projects: the case study of the Chequered Skipper in England. J Insect Conserv 23:419–431. https://doi.org/10.1007/s10841-019-00154-w
Merow C, Smith MJ, Silander JA (2013) A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 36:1058–1069. https://doi.org/10.1111/j.1600-0587.2013.07872.x
Moore JL (2004) The ecology and re-introduction of the chequered skipper butterfly Carterocephalus palaemon in England. PhD thesis, University of Birmingham
Oates M, Warren M (1990) A review of butterfly introductions in Britain and Ireland. Joint Committee for the Conservation of British Insects/World Wildlife Fund, Godalming
Osborne PE, Seddon PJ (2012) Selecting suitable habitats for reintroductions: variation, change and the role of species distribution modelling. Reintroduction Biology: integrating Science and Management. John Wiley and Sons, pp 73–104. https://doi.org/10.1002/9781444355833.ch3
Rosina K, Batista e Silva F, Vizcaino P, Marín Herrera M, Freire S, Schiavina M (2018) Int J Digit Earth 1–25. https://doi.org/10.1080/17538947.2018.1550119. Increasing the detail of European land use/cover data by combining heterogeneous data sets
Shabani F, Kumar L, Ahmadi M (2016) A comparison of absolute performance of different correlative and mechanistic species distribution models in an independent area. Ecol Evol 6:5973–5986. https://doi.org/10.1002/ece3.2332
Smeraldo S, Di Febbraro M, Ćirović D, Bosso L, Trbojević I, Russo D (2017) Species distribution models as a tool to predict range expansion after reintroduction: a case study on eurasian beavers (Castor fiber). J Nat Conserv 37:12–20. https://doi.org/10.1016/j.jnc.2017.02.008
Taylor G, Canessa S, Clarke RH, Ingwersen D, Armstrong DP, Seddon PJ, Ewen JG (2017) Is Reintroduction Biology an Effective Applied Science? Trends Ecol Evol 32:873–880. https://doi.org/10.1016/j.tree.2017.08.002
Telenius A (2011) Biodiversity information goes public: GBIF at your service. Nord J Bot 29:378–381. https://doi.org/10.1111/j.1756-1051.2011.01167.x
Thomas JA, Bourn NAD, Clarke RT, Stewart KE, Simcox DJ, Pearman GS, Curtis R, Goodger B (2001) The quality and isolation of habitat patches both determine where butterflies persist in fragmented landscapes. Proc R Soc Lond B Biol Sci 268:1791–1796. https://doi.org/10.1098/rspb.2001.1693
Thomas JA, Simcox DJ, Clarke RT (2009) Successful conservation of a threatened Maculinea butterfly. Science 325:80–83. https://doi.org/10.1126/science.1175726
Urbanek S (2021) rJava: Low-Level R to Java Interface. R package version 1.0–4
Velazco SJE, Rose MB, De Marco P, Regan HM, Franklin J (2023) How far can I extrapolate my species distribution model? Exploring shape, a novel method. https://doi.org/10.1111/ecog.06992. Ecography e06992
Villero D, Pla M, Camps D, Ruiz-Olmo J, Brotons L (2017) Integrating species distribution modelling into decision-making to inform conservation actions. Biodivers Conserv 26:251–271. https://doi.org/10.1007/s10531-016-1243-2
Vollering Julien, Halvorsen R, Mazzoni S (2019) The MIAmaxent R package: variable transformation and model selection for species distribution models. Ecol Evol 9:12051–12068. https://doi.org/10.1002/ece3.5654
Warren DL, Matzke NJ, Iglesias TL (2020) Evaluating presence-only species distribution models with discrimination accuracy is uninformative for many applications. J Biogeogr 47:167–180. https://doi.org/10.1111/jbi.13705
Weidemann H-J (1988) Tagfalter (Band 2): Biologie, Ökologie, Biotopschutz. Neuman-Neudamm, Germany
Yackulic CB, Chandler R, Zipkin EF, Royle JA, Nichols JD, Grant C, Veran EH, S (2013) Presence-only modelling using MAXENT: when can we trust the inferences? Methods Ecol Evol 4:236–243. https://doi.org/10.1111/2041-210x.12004
Acknowledgements
GH is funded by the NERC ACCE Doctoral training partnership (NERC Grant number NE/S00713X/1) and by Butterfly Conservation. We thank Phillippe Goffart for supporting the project, discussing the ecology and distribution of C. palaemon in Belgium, and facilitating access to the independent validation data set. This dataset was kindly provided by the Public Service of Wallonia free of charge (Origin of the information: SPW/DEMNA and collaborators). The European Butterfly Monitoring Scheme (eBMS) is indebted to the constituent National BMS, their funders and all colleagues (mostly volunteers) who contribute data. The monitoring schemes which contributed data here are funded by Natural Environment Research Council (acting through Centre for Ecology & Hydrology (CEH)), Butterfly Conservation UK, Helmholtz-Zentrum für Umweltforschung GmbH – UFZ, De Vlinderstichting, Butterfly Conservation Europe (BCE), Research Institute Nature and Forest (INBO), Muséum National d’Histoire Naturelle (MNHN) CNRS-UPMC. The Dutch BMS is a co-operation between Dutch Butterfly Conservation and Statistics Netherlands (CBS), part of the Network Ecological Monitoring (NEM) and financed by the Ministry of Agriculture, Nature and Food Quality (LNV). The German BMS is a cooperation between the Helmholtz Centre for Environmental Research - UFZ, German Butterfly Conservation (GfS) and science4you. The French BMS is a cooperation between the Muséum National d’Histoire Naturelle (MNHN), Office Français de la Biodiversité (OFB) et Office pour les Insectes et leur Environnement (OPIE).
Funding
GH is funded by the NERC ACCE Doctoral training partnership (NERC Grant number NE/S00713X/1) and by Butterfly Conservation.
Author information
Authors and Affiliations
Contributions
GH, JAH, CRB and NB conceived and designed the research. AH and DM oversaw data collection in their respective countries. Data analysis was performed by GH and JAH. The first draft of the manuscript was written by GH. All authors commented on subsequent versions and read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
Not applicable.
Competing interests
Dirk Maes is an editor for the Journal of Insect Conservation. Nigel Bourn is a guest editor on a special issue of the journal in 2023. However, neither of these authors will have a role in the editorial decisions for this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Halford, G., Bulman, C.R., Bourn, N. et al. Can species distribution models using remotely sensed variables inform reintroductions? Trialling methods with Carterocephalus palaemon the Chequered Skipper Butterfly. J Insect Conserv (2024). https://doi.org/10.1007/s10841-024-00555-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10841-024-00555-6