Abstract
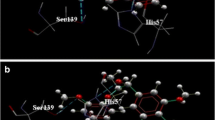
The Hepatitis C Virus (HCV) NS3/4A is an attractive target for the treatment of Hepatitis C infection. Herein, we present an investigation of HCV NS3/4A inhibitors based on a sulfonamidobenzamide scaffold. Inhibitor interactions with HCV NS3/4A were explored by molecular docking, molecular dynamics simulations, and MM/PBSA binding free energy calculations. All of the inhibitors adopt similar molecular docking poses in the catalytic site of the protease that are stabilized by hydrogen bond interactions with G137 and the catalytic S139, which are known to be important for potency and binding stability. The quantitative assessments of binding free energies from MM/PBSA correlate well with the experimental results, with a high coefficient of determination, R2 of 0.92. Binding free energy decomposition analyses elucidate the different contributions of Q41, F43, H57, R109, K136, G137, S138, S139, A156, M485, and Q526 in binding different inhibitors. The importance of these sidechain contributions was further confirmed by computational alanine scanning mutagenesis. In addition, the sidechains of K136 and S139 show crucial but distinct contributions to inhibitor binding with HCV NS3/4A. The structural basis of the potency has been elucidated, demonstrating the importance of the R155 sidechain conformation. This extensive exploration of binding energies and interactions between these compounds and HCV NS3/4A at the atomic level should benefit future antiviral drug design.








Similar content being viewed by others
References
Barathan M, Mohamed R, Yong YK et al (2018) Viral persistence and chronicity in hepatitis C virus infection: role of T-cell apoptosis,senescence and exhaustion. Cells 7:E165. https://doi.org/10.3390/cells7100165
Khadempour-Arani H, Shojaeian A, Mehri-Ghahfarrokhi A et al (2020) Identifying genotype profile of chronic hepatitis C infection in Southwest Iran. J Res Med Sci 25:85. https://doi.org/10.4103/jrms.JRMS_524_19
Aishanjiang K, Wei X, Fu Y et al (2021) Circular RNAs and hepatocellular carcinoma: new epigenetic players with diagnostic and prognostic roles. Front Oncol. https://doi.org/10.3389/fonc.2021.653717
Frazzoni L, Sikandar U, Metelli F et al (2021) Hepatocellular carcinoma recurrence after hepatitis C virus therapy with direct-acting antivirals. A systematic review and meta-analysis. J Clin Med 10:1694. https://doi.org/10.3390/jcm10081694
Teng W, Liu YC, Jeng WJ, Su CW (2021) Tertiary prevention of HCC in chronic hepatitis B or C infected patients. Cancers Basel 13:1729. https://doi.org/10.3390/cancers13071729
Smith DB, Bukh J, Kuiken C et al (2014) Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource. Hepatology 59:318–327. https://doi.org/10.1002/hep.26744
Kim CW, Chang K-M (2013) Hepatitis C virus: virology and life cycle. Clin Mol Hepatol 19:17–25. https://doi.org/10.3350/cmh.2013.19.1.17
Lohmann V, Korner F, Koch J-O et al (1999) Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113
Paul D, Madan V, Bartenschlager R (2014) Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe 16:569–579
Kim JL, Morgenstern KA, Lin C et al (1996) Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell 87:343–355. https://doi.org/10.1016/S0092-8674(00)81351-3
Love RA, Parge HE, Wickersham JA et al (1996) The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell 87:331–342
Tran HTL, Morikawa K, Anggakusuma, et al (2020) OCIAD1 is a host mitochondrial substrate of the hepatitis C virus NS3-4A protease. PLoS ONE 15:e0236447. https://doi.org/10.1371/journal.pone.0236447
Moradpour D, Penin F (2013) Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol 369:113–142. https://doi.org/10.1007/978-3-642-27340-7_5
Jiang Y, Andrews SW, Condroski KR et al (2014) Discovery of danoprevir (ITMN-191/R7227), a highly selective and potent inhibitor of hepatitis C virus (HCV) NS3/4A protease. J Med Chem 57:1753–1769
Kwong AD, Kauffman RS, Hurter P, Mueller P (2011) Discovery and development of telaprevir: an NS3-4A protease inhibitor for treating genotype 1 chronic hepatitis C virus. Nat Biotechnol 29:993–1003
Nageswara Rao D, Zephyr J, Henes M et al (2021) Discovery of quinoxaline-based p1–p3 macrocyclic ns3/4a protease inhibitors with potent activity against drug-resistant hepatitis C virus variants. J Med Chem 64:11972–11989. https://doi.org/10.1021/acs.jmedchem.1c00554
Neelamkavil SF, Agrawal S, Bara T et al (2016) Discovery of MK-8831, a novel spiro-proline macrocycle as a pan-genotypic HCV-NS3/4a protease inhibitor. ACS Med Chem Lett 7:111–116
Rosenquist Å, Samuelsson B, Johansson P-O et al (2014) Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor. J Med Chem 57:1673–1693
Shah U, Jayne C, Chackalamannil S et al (2014) Novel quinoline-based P2–P4 macrocyclic derivatives as pan-genotypic HCV NS3/4a protease inhibitors. ACS Med Chem Lett 5:264–269
Summa V, Ludmerer SW, McCauley JA et al (2012) MK-5172, a selective inhibitor of hepatitis C virus NS3/4a protease with broad activity across genotypes and resistant variants. Antimicrob Agents Chemother 56:4161–4167
Gentile I, Buonomo AR, Zappulo E et al (2014) Asunaprevir, a protease inhibitor for the treatment of hepatitis C infection. Ther Clin Risk Manag. 10:493–504. https://doi.org/10.2147/TCRM.S66731
McCauley JA, McIntyre CJ, Rudd MT et al (2010) Discovery of vaniprevir (MK-7009), a macrocyclic hepatitis C virus NS3/4a protease inhibitor. J Med Chem 53:2443–2463. https://doi.org/10.1021/jm9015526
Agarwal A, Zhang B, Olek E et al (2012) Rapid and sharp decline in HCV upon monotherapy with NS3 protease inhibitor, ACH-1625. Antivir Ther 17:1533–1539. https://doi.org/10.3851/Imp2359
Sheng XC, Appleby T, Butler T et al (2012) Discovery of GS-9451: an acid inhibitor of the hepatitis C virus NS3/4A protease. Bioorg Med Chem Lett 22:2629–2634
Li H, Scott JP, Chen C et al (2015) Synthesis of bis-macrocyclic HCV protease inhibitor MK-6325 via intramolecular sp2–sp3 Suzuki-Miyaura coupling and ring closing metathesis. Org Lett 17:1533–1536. https://doi.org/10.1021/acs.orglett.5b00418
Butt AA, Kanwal F (2012) Boceprevir and telaprevir in the management of hepatitis C virus-infected patients. Clin Infect Dis 54:96–104. https://doi.org/10.1093/cid/cir774
Gentile I, Borgia F, Riccardo Buonomo A et al (2014) ABT-450: a novel protease inhibitor for the treatment of hepatitis C virus infection. Curr Med Chem 21:3261–3270
Lawitz EJ, O’Riordan WD, Asatryan A et al (2016) Potent antiviral activities of the direct-acting antivirals ABT-493 and ABT-530 with three-day monotherapy for hepatitis C virus genotype 1 infection. Antimicrob Agents Chemother 60:1546–1555
Poordad F, Dieterich D (2012) Treating hepatitis C: current standard of care and emerging direct-acting antiviral agents. J Viral Hepat 19:449–464. https://doi.org/10.1111/j.1365-2893.2012.01617.x
Classification of Direct-Acting Antiviral Agents in HCV Treatment Regimens. https://hepatitis.va.gov/hcv/treatment/hcv-daa-class.asp
Amoako A, Ortiz-Paredes D, Engler K et al (2021) Patient and provider perceived barriers and facilitators to direct acting antiviral hepatitis C treatment among priority populations in high income countries: A knowledge synthesis. Int J Drug Policy 96:103247. https://doi.org/10.1016/j.drugpo.2021.103247
Danilescu CM, Ionescu M, Sandulescu DL et al (2022) Perceived stress in hepatitis C Virus infected patients under the DAA-based therapy. Diagnostics 12:1177. https://doi.org/10.3390/diagnostics12051177
Roger S, Ducancelle A, Le Guillou-Guillemette H et al (2021) HCV virology and diagnosis. Clin Res Hepatol Gastroenterol 45:101626
Velazquez-Moctezuma R, Augestad EH, Castelli M et al (2021) Mechanisms of hepatitis C virus escape from vaccine-relevant neutralizing antibodies. Vaccines Basel 9:291. https://doi.org/10.3390/vaccines9030291
Jin G, Lee J, Lee K (2017) Chemical genetics-based development of small molecules targeting hepatitis C virus. Arch Pharm Res 40:1021–1036. https://doi.org/10.1007/s12272-017-0949-3
Forton DM (2016) How much of a problem is resistance in treating hepatitis C? Curr Opin Infect Dis 29:625–631. https://doi.org/10.1097/QCO.0000000000000319
Shoun AA, Abozahra R, Baraka K et al (2022) Identifying different mutation sites leading to resistance to the Direct-Acting Antiviral (DAA) sofosbuvir in hepatitis C virus patients from Egypt. Microorganisms 10:679. https://doi.org/10.3390/microorganisms10040679
Park SB, Boyer A, Hu Z et al (2019) Discovery and characterization of a novel HCV inhibitor targeting the late stage of HCV life cycle. Antivir Ther 24:371–381. https://doi.org/10.3851/IMP3303
Johansson A, Poliakov A, Åkerblom E et al (2003) Acyl sulfonamides as potent protease inhibitors of the hepatitis C virus full-Length NS3 (Protease-Helicase/NTPase): A comparative study of different C-terminals. Bioorg Med Chem 11:2551–2568. https://doi.org/10.1016/S0968-0896(03)00179-2
LaPlante SR, Nar H, Lemke CT et al (2014) Ligand bioactive conformation plays a critical role in the design of drugs that target the hepatitis C virus NS3 protease. J Med Chem 57:1777–1789
Rönn R, Sabnis YA, Gossas T et al (2006) Exploration of acyl sulfonamides as carboxylic acid replacements in protease inhibitors of the hepatitis C virus full-length NS3. Bioorg Med Chem 14:544–559. https://doi.org/10.1016/j.bmc.2005.08.045
Chaudhuri R, Lee H, Truong L et al (2012) Identification of non-macrocyclic small molecule inhibitors against the NS3/4A Serine protease of hepatitis C virus through in silico screening. J Chem Inf Model 52:2245–2256. https://doi.org/10.1021/ci300177p
Lee H, Zhu T, Patel K et al (2013) High-throughput screening (HTS) and hit validation to identify small molecule inhibitors with activity against NS3/4A proteases from multiple hepatitis C virus genotypes. PLoS ONE 8:e75144
Ren J, Ojeda I, Patel M et al (2019) Exploring small molecules with pan-genotypic inhibitory activities against hepatitis C virus NS3/4A serine protease. Bioorg Med Chem Lett 29:2349–2353. https://doi.org/10.1016/j.bmcl.2019.06.009
Vaid TM, Chalmers DK, Scott DJ, Gooley PR (2020) INPHARMA-based determination of ligand binding modes at α1 -adrenergic receptors explains the molecular basis of subtype selectivity. Chem Weinh Bergstr Ger 26:11796–11805. https://doi.org/10.1002/chem.202000642
Ejeh S, Uzairu A, Shallangwa GA et al (2022) In silico design and pharmacokinetics investigation of some novel hepatitis C virus NS5B inhibitors: pharmacoinformatics approach. Bull Natl Res Cent 46:109. https://doi.org/10.1186/s42269-022-00796-y
Friedman R (2022) Computational studies of protein–drug binding affinity changes upon mutations in the drug target. WIREs Comput Mol Sci 12:e1563. https://doi.org/10.1002/wcms.1563
Hdoufane I, Bjij I, Oubahmane M et al (2022) In silico design and analysis of NS4B inhibitors against hepatitis C virus. J Biomol Struct Dyn 40:1915–1929. https://doi.org/10.1080/07391102.2020.1839561
Hussain R, Khalid H, Fatmi MQ (2022) HCV genotype-specific drug discovery through structure-based virtual screening. Pure Appl Chem 94:809–818. https://doi.org/10.1515/pac-2021-1104
Sahakyan H (2021) Improving virtual screening results with MM/GBSA and MM/PBSA rescoring. J Comput Aided Mol Des 35:731–736. https://doi.org/10.1007/s10822-021-00389-3
Su PC, Tsai CC, Mehboob S et al (2015) Comparison of radii sets, entropy, QM methods, and sampling on MM-PBSA, MM-GBSA, and QM/MM-GBSA ligand binding energies of F-tularensis enoyl-ACP reductase (FabI). J Comput Chem 36:1859–1873. https://doi.org/10.1002/jcc.24011
Zhu Y-X, Sheng Y-J, Ma Y-Q, Ding H-M (2022) Assessing the performance of screening MM/PBSA in Protein-ligand interactions. J Phys Chem B 126:1700–1708. https://doi.org/10.1021/acs.jpcb.1c09424
Schiering N, D’Arcy A, Villard F et al (2011) A macrocyclic HCV NS3/4A protease inhibitor interacts with protease and helicase residues in the complex with its full-length target. Proc Natl Acad Sci 108:21052–21056
Korb O, Stützle T, Exner TE (2009) Empirical scoring functions for advanced protein−ligand docking with plants. J Chem Inf Model 49:84–96. https://doi.org/10.1021/ci800298z
Verdonk ML, Cole JC, Hartshorn MJ et al (2003) Improved protein–ligand docking using GOLD. Proteins Struct Funct Bioinforma 52:609–623
Cheng HC (2001) The power issue: determination of KB or Ki from IC50: a closer look at the Cheng-Prusoff equation, the Schild plot and related power equations. J Pharmacol Toxicol Methods 46:61–71. https://doi.org/10.1016/S1056-8719(02)00166-1
Feig M, Onufriev A, Lee Michael S et al (2004) Performance comparison of generalized born and Poisson methods in the calculation of electrostatic solvation energies for protein structures. J Comput Chem 25:265–284. https://doi.org/10.1002/jcc.10378
Romano KP, Ali A, Royer WE, Schiffer CA (2010) Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc Natl Acad Sci U A 107:20986–20991. https://doi.org/10.1073/pnas.1006370107
Taylor JG, Zipfel S, Ramey K et al (2019) Discovery of the pan-genotypic hepatitis C virus NS3/4A protease inhibitor voxilaprevir (GS-9857): a component of Vosevi®. Bioorg Med Chem Lett 29:2428–2436. https://doi.org/10.1016/j.bmcl.2019.03.037
Yi M, Tong X, Skelton A et al (2006) Mutations conferring resistance to SCH6, a novel hepatitis C virus NS3/4A protease inhibitor. Reduced RNA replication fitness and partial rescue by second-site mutations. J Biol Chem 281:8205–8215. https://doi.org/10.1074/jbc.M510246200
Arasappan A, Bennett F, Bogen SL et al (2021) RCSB PDB - 3LON: HCV NS3-4a protease domain with ketoamide inhibitor narlaprevir. www.rcsb.org/structure/3LON. Accessed 1 Aug 2022
Bennett F, Huang Y, Hendrata S et al (2010) The introduction of P4 substituted 1-methylcyclohexyl groups into Boceprevir: a change in direction in the search for a second generation HCV NS3 protease inhibitor. Bioorg Med Chem Lett 20:2617–2621. https://doi.org/10.1016/j.bmcl.2010.02.063
Heyda J, Mason PE, Jungwirth P (2010) Attractive interactions between side chains of histidine-histidine and histidine-arginine-based cationic dipeptides in water. J Phys Chem B 114:8744–8749. https://doi.org/10.1021/jp101031v
Lee H, Torres J, Truong L et al (2012) Reducing agents affect inhibitory activities of compounds: results from multiple drug targets. Anal Biochem 423:46–53
Sastry GM, Adzhigirey M, Day T et al (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/s10822-013-9644-8
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS All-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc 118:11225–11236. https://doi.org/10.1021/ja9621760
Schrödinger L (2022) Schrödinger Release 2022–2: LigPrep. Schrödinger, LLC, New York
Pettersen EF, Goddard TD, Huang CC et al (2004) UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Abagyan R, Totrov M, Kuznetsov D (1994) ICM—A new method for protein modeling and design: applications to docking and structure prediction from the distorted native conformation. J Comput Chem 15:488–506. https://doi.org/10.1002/jcc.540150503
Case DA, Babin V, Berryman J, et al (2014) Amber 14
Vanquelef E, Simon S, Marquant G et al (2011) R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res 39:W511–W517. https://doi.org/10.1093/nar/gkr288
Wang JM, Wolf RM, Caldwell JW et al (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174. https://doi.org/10.1002/jcc.20035
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. https://doi.org/10.1063/1.464397
Kräutler V, van Gunsteren WF, Hünenberger PH (2001) A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem 22:501–508. https://doi.org/10.1002/1096-987X(20010415)22:5%3c501::AID-JCC1021%3e3.0.CO;2-V
Roe DR, Cheatham TE III (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9:3084–3095
Duan L, Feng G, Wang X et al (2017) Effect of electrostatic polarization and bridging water on CDK2–ligand binding affinities calculated using a highly efficient interaction entropy method. Phys Chem Chem Phys 19:10140–10152
Duan L, Liu X, Zhang JZ (2016) Interaction entropy: a new paradigm for highly efficient and reliable computation of protein–ligand binding free energy. J Am Chem Soc 138:5722–5728
Duan LL, Feng GQ, Zhang QG (2016) Large-scale molecular dynamics simulation: effect of polarization on thrombin-ligand binding energy. Sci Rep 6:1–11
Acknowledgements
We thank ChemAxon for a free academic license of their cheminformatics suite, used for data management. This work was supported in part by NIH grant R41 AI112114. Molecular graphics and analyses were performed with UCSF Chimera (developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311). Access to ICM-Molsoft (version 3.9-2e) was generously provided by Proteco Research LLC. The MD simulations were performed on the UIC Extreme high-performance computing cluster in the UIC Academic Computing and Communications Center. We thank UIC ACCC providing us with access.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design, including material preparation, data collection and analysis. The initial draft of the manuscript was written by JR, with additions and revisions from TMV. HL and MEJ provided comments and edited the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests related to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ren, J., Vaid, T.M., Lee, H. et al. Evaluation of interactions between the hepatitis C virus NS3/4A and sulfonamidobenzamide based molecules using molecular docking, molecular dynamics simulations and binding free energy calculations. J Comput Aided Mol Des 37, 53–65 (2023). https://doi.org/10.1007/s10822-022-00490-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-022-00490-1