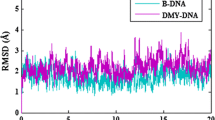
Abstract
DNA is an essential target for the treatment of various pathologies, especially cancer. Hence targeting DNA double helix for alteration of its function has been attempted by several ways. Drug–DNA intercalation, one such biophysical process, could not be studied extensively as this requires significant deformation of the receptor DNA. Here we report thorough theoretical investigation of intercalation process in daunomycin–DNA interaction, by performing molecular dynamics simulations of the drug–DNA complexes for various DNA sequences, followed by Free-energy analysis and density functional theory (DFT) based studies to understand the binding preference. The classical energy based analyses indicate that the drug prefers to bind to TC/GA sequence over others. The DFT based energies of supra-molecular complexes are always contaminated with basis set superposition error (BSSE), which can be corrected by counterpoise method. This method is quite effective for systems containing two molecular fragments but is not appropriate for studying interaction between two base pair fragments and the drug intercalated between them. We have adopted an extension of the counterpoise method for BSSE corrected interaction energy calculation. These interaction energies, along with the energy penalty due to un-stacking of the base pairs, also indicate TC/GA sequence is the most preferred sequence for binding.





Similar content being viewed by others
References
Gewirtz DA (1999) Biochem Pharmacol 57:727–741
Ghosh D, Hossain M, Saha C, Dey SK, Kumar GS (2012) DNA Cell Biol 31:378–387
Da Ros T, Spalluto G, Prato M, Saison-Behmoaras T, Boutorine A, Cacciari B (2005) Curr Med Chem 12:71–88
Pabo CO, Sauer RT (1984) Annu Rev Biochem 53:293–321
Gilad Y, Senderowitz H (2014) J Chem Inf Model 54:96–107
Chaires JB (1998) Curr Opin Struct Biol 8:314–320
Ihmels LTH (2011) Intercalation of organic ligands as a tool to modify the properties of DNA. In: Jin J-I, Grote J (eds) Materials science of DNA. CRC Press, Boca Raton
Halder S, Bhattacharyya D (2013) Prog Biophys Mol Biol 113:264–283
Chaires JB, Fox KR, Herrera JE, Britt M, Waring MJ (1987) Biochemistry 26:8227–8236
Chaires JB, Herrera JE, Waring MJ (1990) Biochemistry-Us 29:6145–6153
Chen KX, Gresh N, Pullman B (1985) J Biomol Struct Dyn 3:445–466
Hurley LH (2002) Nat Rev Cancer 2:188–200
Todd AK, Adams A, Thorpe JH, Denny WA, Wakelin LPG, Cardin CJ (1999) J Med Chem 42:536–540
Yeh HJC, Sayer JM, Liu XH, Altieri AS, Byrd RA, Lakshman MK, Yagi H, Schurter EJ, Gorenstein DG, Jerina DM (1995) Biochemistry 34:13570–13581
Boer DR, Canals A, Coll M (2009) Dalton Trans 3:399–414
Mukherjee A (2011) J Phys Chem Lett 2:3021–3026
Mukherjee A, Lavery R, Bagchi B, Hynes JT (2008) J Am Chem Soc 130:9747–9755
Wilhelm M, Mukherjee A, Bouvier B, Zakrzewska K, Hynes JT, Lavery R (2012) J Am Chem Soc 134:8588–8596
Baginski M, Fogolari F, Briggs JM (1997) J Mol Biol 274:253–267
Rehn C, Pindur U (1996) Monatsh Chem 127:631–644
Bailly C, Echepare S, Gago F, Waring MJ (1999) Anti-Cancer Drug Des 14:291–303
Chen KX, Gresh N, Pullman B (1986) Mol Pharmacol 30:279–286
Medhi C, Mitchell JBO, Price SL, Tabor AB (1999) Biopolymers 52:84–93
Hannon MJ (2007) Chem Soc Rev 36:280–295
Wheate NJ, Brodie CR, Collins JG, Kemp S, Aldrich-Wright JR (2007) Mini Rev Med Chem 7:627–648
Mondal M, Mukherjee S, Bhattacharyya D (2014) J Mol Model 20:2499
Bell CE, Lewis M (2000) Nat Struct Biol 7:209–214
Romanuka J, Folkers GE, Biris N, Tishchenko E, Wienk H, Bonvin AM, Kaptein R, Boelens R (2009) J Mol Biol 390:478–489
Werner MH, Gronenborn AM, Clore GM (1996) Science 271:778–784
Firczuk M, Wojciechowski M, Czapinska H, Bochtler M (2011) Nucleic Acids Res 39:744–754
Sandmann A, Sticht H (2018) PLoS ONE 13:e0192605
Goerigk L, Grimme S (2011) Phys Chem Chem Phys 13:6670–6688
Hohenstein EG, Chill ST, Sherrill CD (2008) J Chem Theory Comput 4:1996–2000
Zhao Y, Truhlar DG (2008) Accounts Chem Res 41:157–167
Chai JD, Head-Gordon M (2008) Phys Chem Chem Phys 10:6615–6620
Arago J, Sancho-Garcia JC, Orti E, Beljonne D (2011) J Chem Theory Comput 7:2068–2077
Dunning TH (1989) J Chem Phys 90:1007–1023
Peterson KA, Kendall RA, Dunning TH (1993) J Chem Phys 99:1930–1944
Morgado C, Vincent MA, Hillier IH, Shan X (2007) Phys Chem Chem Phys 9:448–451
Reha D, Kabelac M, Ryjacek F, Sponer J, Sponer JE, Elstner M, Suhai S, Hobza P (2002) J Am Chem Soc 124:3366–3376
Morokuma K (1971) J Chem Phys 55:1236–2000
Boys SF, Bernardi F (2002) Mol Phys 100:65–73
Richard RM, Bakr BW, Sherrill CD (2018) J Chem Theory Comput 14:2386–2400
Phipps MJS, Fox T, Tautermann CS, Skylaris CK (2015) Chem Soc Rev 44:3177–3211
Turney JM, Simmonett AC, Parrish RM, Hohenstein EG, Evangelista FA, Fermann JT, Mintz BJ, Burns LA, Wilke JJ, Abrams ML, Russ NJ, Leininger ML, Janssen CL, Seidl ET, Allen WD, Schaefer HF, King RA, Valeev EF, Sherrill CD, Crawford TD (2012) WIREs Comput Mol Sci 2:556–565
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Res 28:235–242
Wang AHJ, Ughetto G, Quigley GJ, Rich A (1987) Biochemistry 26:1152–1163
Bhattacharyya D, Halder S, Basu S, Mukherjee D, Kumar P, Bansal M (2017) J Comput Aid Mol Des 31:219–235
Chandrasekaran R, Arnott S (1996) J Biomol Struct Dyn 13:1015–1027
Bansal M, Bhattacharyya D, Ravi B (1995) Comput Appl Biosci 11:281–287
Pingali PK, Halder S, Mukherjee D, Basu S, Banerjee R, Choudhury D, Bhattacharyya D (2014) J Comput Aid Mol Des 28:851–867
Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M (2009) J Comput Chem 30:1545–1614
DeLano WL (2002) CCP4 Newslett Protein Crystallogr 40:82–92
Galindo-Murillo R, Robertson JC, Zgarbova M, Sponer J, Otyepka M, Jurecka P, Cheatham TE (2016) J Chem Theory Comput 12:4114–4127
Besler BH, Merz KM, Kollman PA (1990) J Comput Chem 11:431–439
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) J Am Chem Soc 117:5179–5197
Frisch GWTMJ, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA Jr, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA (2003) Gaussian 09, Revision B.01. Gaussian Inc, Pittsburgh
Cornell WD, Cieplak P, Bayly CI, Kollman PA (1993) J Am Chem Soc 115:9620–9631
Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25:1157–1174
Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) J Comput Chem 26:1668–1688
Mobley DL, Chodera JD, Dill KA (2006) J Chem Phys 125:084902
Mark P, Nilsson L (2001) J Phys Chem A 105:9954–9960
Press WHT, Vetterling SA, Flannery BP (1993) Numeric recipes: the art of scientific computing, 3rd edn. Cambridge University Press, New York
Hess B, Bekker H, Berendsen HJC, Fraaije JGEM (1997) J Comput Chem 18:1463–1472
Parrinello M, Rahman A (1981) J Appl Phys 52:7182–7190
Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR (1984) J Chem Phys 81:3684–3690
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089–10092
Mukherjee S, Bansal M, Bhattacharyya D (2006) J Comput Aid Mol Des 20:629–645
Barone G, Guerra CF, Bickelhaupt FM (2013) Chemistryopen 2:186–193
Mukherjee S, Kailasam S, Bansal M, Bhattacharyya D (2014) Biopolymers 101:107–120
Svozil D, Hobza P, Sponer J (2010) J Phys Chem B 114:2547–2547
Frisch GWTMJ, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Haseg J, Fox DJ (2010) Guassian 09, revision A02. Gaussian Inc., Wallingford
Barone V, Cossi M (1998) J Phys Chem A 102:1995–2001
Miller BR, McGee TD, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) J Chem Theory Comput 8:3314–3321
Samanta S, Mukherjee S, Chakrabarti J, Bhattacharyya D (2009) J Chem Phys 130:03B614
Calladine CR (1982) J Mol Biol 161:343–352
Duarte CM, Pyle AM (1998) J Mol Biol 284:1465–1478
Brown TN, Mora-Diez N (2006) J Phys Chem B 110:9270–9279
East ALL, Smith BJ, Radom L (1997) J Am Chem Soc 119:9014–9020
Halder A, Halder S, Bhattacharyya D, Mitra A (2014) Phys Chem Chem Phys 16:18383–18396
Klamt A, Eckert F, Diedenhofen M, Beck ME (2003) J Phys Chem A 107:9380–9386
Munegumi T (2013) World J Chem Educ 1:12–16
Serjeant AAP (1984) The determination of ionization constant. Chapman and Hall, London
Li L, Li CA, Sarkar S, Zhang J, Witham S, Zhang Z, Wang L, Smith N, Petukh M, Alexov E (2012) BMC Biophys 5:9
Morgado CA, Svozil D, Turner DH, Sponer J (2012) Phys Chem Chem Phys 14:12580–12591
Basham B, Schroth GP, Ho PS (1995) Proc Natl Acad Sci USA 92:6464–6468
Chakrabarti S, Bhattacharyya D, Dasgupta D (2000) Biopolymers 56:85–95
Beveridge DL, Barreiro G, Byun KS, Case DA, Cheatham TE, Dixit SB, Giudice E, Lankas F, Lavery R, Maddocks JH, Osman R, Seibert E, Sklenar H, Stoll G, Thayer KM, Varnai P, Young MA (2004) Biophys J 87:3799–3813
Acknowledgements
The authors thank Prof. Ashoke Prasun Chattopadhyay for useful discussions. L.M thanks SERB-DST, Govt. of India for providing financial assistant under NPDF project SERB/F/85I2/20I7-20I8. Authors thank DAE, Govt. of India, under CAPP-II project and BRAF facility of CDAC, Pune, India for computational facility.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Maganti, L., Bhattacharyya, D. Sequence specificity in DNA–drug intercalation: MD simulation and density functional theory approaches. J Comput Aided Mol Des 34, 83–95 (2020). https://doi.org/10.1007/s10822-019-00268-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00268-y