Abstract
Stabilizing unique receptor conformations, allosteric modulators of G-protein coupled receptors (GPCRs) might open novel treatment options due to their new pharmacological action, their enhanced specificity and selectivity in both binding and signaling. Ligand binding occurs at intrahelical allosteric sites and involves significant induced fit effects that include conformational changes in the local protein environment and water networks. Based on the analysis of available crystal structures of metabotropic glutamate receptor 5 (mGlu5) we investigated these effects in the binding of mGlu5 receptor negative allosteric modulators. A large set of retrospective virtual screens revealed that the use of multiple protein structures and the inclusion of selected water molecules improves virtual screening performance compared to conventional docking strategies. The role of water molecules and protein flexibility in ligand binding can be taken into account efficiently by the proposed docking protocol that provided reasonable enrichment of true positives. This protocol is expected to be useful also for identifying intrahelical allosteric modulators for other GPCR targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Structure-based virtual screening is an established computational technique for the identification of chemical starting points in drug discovery programs [1]. Virtual screening allows to select compounds for experimental testing that improves the efficiency of hit finding. Structure-based screening requires atomic resolution protein structures that are available either from experiment or as computational models. Owing to the increasing availability of GPCR X-ray structures these important class of proteins are also extensively and successfully used in structure-based virtual screening campaigns [2]. Typically, GPCR virtual screen aims to identify ligands binding in the intrahelical orthosteric binding site.
GPCRs contain a wide variety of allosteric sites and their targeting with small molecules offers unique therapeutic potential [3,4,5]. As it has been discussed recently [6] allosteric sites are widely distributed intrahelically in the TM bundle, at extrahelical positions within the membrane binding region and at the intracellular signalling protein interface. These sites may or may not be preformed before ligand binding and they are generally sensitive to protein conformational changes and to the presence of and the interactions with water molecules. We selected the metabotropic glutamate receptor 5 (mGlu5 receptor) for a detailed investigation because its allosteric site is structurally well characterized owing to the receptor complexes co-crystallized with a variety of allosteric ligands [7,8,9,10]. Moreover, a wide range of allosteric ligands with biochemical data have been reported. Therefore, the mGlu5 receptor is well suited to investigate how virtual screening can find allosteric ligands and what are the special considerations needed for maximizing its value in allosteric ligand discovery. Metabotropic glutamate receptors belong to the family C of GPCRs, and can influence neurotransmitter release or modulate ion channel activity [11]. Their endogen ligand is l-glutamic acid, a major excitatory neurotransmitter in the central nervous system (CNS) of mammals. Modulation of glutamate transmission has a potential in the treatment of several psychiatric and neurological disorders. Glutamate binds to the extracellular Venus flytrap domain that is linked to the seven transmembrane domain through a cysteine-rich domain. Non-competitive antagonists which bind in the allosteric site are called negative allosteric modulators (NAMs), and could be useful in the treatment of numerous diseases [12].
Early drug discovery efforts targeted the orthosteric binding site, but this site is highly conserved in the mGlu receptor family, making orthosteric modulation by subtype selective ligands highly challenging. However, selective allosteric modulation with ligands targeting the ancestral intrahelical site (whose position resemble to the orthosteric site of most GPCRs) [13] appears to be more attainable. Large number of allosteric modulators have been reported, although few of them have reached clinic to date [14]. This can be attributed to the significant challenges in the medicinal chemistry optimization of allosteric ligands. The complex, multiparametric approach, characteristic to medicinal chemistry optimizations in general, is further complicated by other factors like allosteric coupling and cooperativity, functional activity and effect on signalling that all have to be taken into account [13].
The allosteric binding site of mGlu5 receptor can be found within the heptahelical transmembrane domain and this pocket is completely covered by the second extracellular loop (EC2) [7]. The transmembrane helices (TM2, TM5 and TM7) of mGlu5 receptor are moved closer to each other and consequently the volume of the pocket is significantly reduced with respect to the typical GPCR class A and class B intrahelical pockets. These latter pockets are the binding sites of their endogenous ligands and they are also the principal target of compounds designed for GPCR activity modulation. In contrast, mGlu5 receptor intrahelical pocket is a functional water channel whose water molecules are likely to participate in signalling [9, 15]. This narrow pocket accommodates a well-structured water network and no small molecule is known to bind for any physiological function. Nevertheless, several exogenous compounds binding to this site have been identified. Their optimization into drug candidates, however, is hindered by irregularly steep structure–activity relationship (SAR), functional switches by small structural changes and limited SAR transferability between chemotypes [14].
X-ray structures of complexes formed between allosteric ligands (Fig. 1) and mGlu5 receptor proved to be indispensable to understand the unconventional features of ligand binding.
The allosteric pocket of mGlu5 receptor is made up of a lower and an upper chamber connected by a narrow linker region. The upper chamber is present throughout the mGlu family and its location is similar to the orthosteric binding pocket of class A GPCRs. However, the lower chamber is present only in mGlu5 receptor owing to the residues Pro6553.40 Gly6282.49 and Ala8107.41, all are smaller than the corresponding residues in the seven other mGlu receptors. In particular, the small size of Pro655 creates a narrow channel towards the lower chamber allowing the ligands to penetrate deeper into the crevice among the helices.
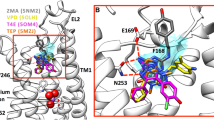
Available complex structures of the mGlu5 receptor contain ligands with three different linkers. Mavoglurant [16] and M-MPEP [17] both have an acetylenic linker, a urea linker is present in fenobam [18] and pyrimidine in HTL-14242 (4) and in 5 [8]. Although mavoglurant and M-MPEP share a common acetylenic linker, this moiety does not perfectly overlay in the two crystal structures and the growing vectors of the headgroups (condensed rings in mavoglurant and methoxyphenyl in M-MPEP) have different orientation for the two ligands [9]. These findings clearly show that modelling of ligand binding is challenging even for a chemotype with available X-ray structures. Indeed, docking of M-MPEP into the protein conformation obtained from the mGlu5 receptor–mavoglurant complex results in two distinct cluster of poses, one that corresponds to the X-ray structure, and another in which the methoxyphenyl group is rotated by 180°. The mGlu5 receptor binding pocket is significantly different in the mavoglurant and the M-MPEP complexes primarily owing to the different conformations adopted by Trp7856.50. The indole ring of Trp7856.50 points outward, toward TM5 in the former (χ1 = − 170°; χ2 = 92°), and it points inward, towards the centre of the helical bundle in the latter (χ1 = − 76°; χ2 = 63°) (Fig. 2). These two conformations of Trp7856.50 result in different shapes of the allosteric binding pocket and warn that protein flexibility and induced fit binding have to be taken into account in the molecular modelling of ligand binding to mGlu5 receptor. This is also supported by several structure-based computational studies which were performed previously to map ligand–protein interactions and to understand irregular SAR using pharmacophore modelling, molecular docking and molecular dynamics simulations [15, 19,20,21,22,23,24,25]. Moreover, analysis of the X-ray structures coupled with computational water network analysis performed by WaterFLAP showed that many of the ligand–protein interactions are mediated by water molecules and this also has to be taken into account in interpreting structure–activity relationships across chemotypes. We found that self-docking of M-MPEP into empty mGlu5 receptor (PDB: 6FFI) results in a pose rotated by 180° with respect to the X-ray pose. In contrast, including the water molecule consistently observed in the X-ray structures results in the X-ray docking pose. This finding confirms that ligand binding is importantly affected by the presence of this water molecule. The sensitivity of the protein conformation, protein–ligand and water mediated interactions to small variations of ligand structures [9] results in unexpected binding modes and irregular structure–activity relationships, and hinders the application of both structure-, and ligand-based modelling methods in a standard manner.
In the present study we evaluate the role of protein flexibility and allosteric binding site water network on structure-based virtual screening on the mGlu5 receptor. Similar analysis was performed for the orthosteric site of class A GPCRs [26,27,28] but the special features of the mGlu5 receptor necessitates a separate analysis for this class C receptor. The intrahelical site of mGlu5 receptor is the ancestral pocket of GPCRs that is the orthosteric site in class A GPCRs, but appears to be a functional water channel involved in the activation process in mGlu receptors. We selected three mGlu5 receptor complexes with available X-ray structures; complexes of mavoglurant, fenobam and HTL-14242 (4). The first two ligands share a linear shape in contrast to ligand 4 having a bent shape (Fig. 1). The other two complexes with available X-ray structures (M-MPEP [9], 5 [8]) were also investigated for including them into the study. M-MPEP is similar to mavoglurant in having acetylenic linker, however, it was observed that this protein structure is able to accommodate less than 40% of the acetylenic compounds compiled for docking. This finding shows that the protein conformation is not only well adapted to the thin acetylenic linker but also to the limited extension of the head groups. Indeed, the methoxyphenyl group of M-MPEP is significantly smaller than the corresponding methyl-octahydroindole-carboxylate group in mavoglurant and this results in the inward orientation of Trp7856.50 and spatially highly restrictive allosteric site. Therefore, the protein structure of the M-MPEP complex was not used in our study. X-ray structure for compound 5 is also available, however, this compound and the corresponding protein structure is highly similar to that of HTL-14242 and the higher resolution structure of the latter (R(5CGC) = 3.1 Å vs. R(5CGD) = 2.6 Å) [8] was selected for our study.
Virtual screening was performed with compound sets of known active ligands and physically similar decoys. Negative allosteric modulators were only considered as active compounds as all available high resolution mGlu5 crystal structures include NAM complexes, and protein conformations in PAM complexes are expected to be significantly different based on experimental [10] and computational analyses [15, 29, 30]. Because of the small number of low affinity NAMs reported for the mGlu5 receptor (503 with IC50 > 10 µM, 429 with IC50 > 5 µM activity) DUD-E [31, 32] was used to generate decoys with physico-chemical properties similar to the actives but with dissimilar fingerprints to minimize topological similarity. Early enrichment factors of protein structures derived from three complexes and containing varying number of water molecules were investigated to explore the effect of protein flexibility and water network on the performance of structure based virtual screening.
Previous docking studies into mGlu receptors explored ligand–protein interactions and their variations depending on receptor subtype [23, 33, 34] and on differences in ligand structures [35] and included the effect of protein flexibility and the interactions with water molecules at various levels. A comprehensive treatment of protein flexibility by induced fit docking and the interactions with explicit water molecules is presented by Harpsøe et al. [23] in a comparative analysis of the allosteric sites of mGlu receptor subtypes and their interactions with various ligands. In contrast, our objective is to devise a virtual screening scheme that takes into account protein flexibility and water mediated interactions found to be crucial in mGlu5 receptor–ligand binding. Conventional docking methods treat the protein environment as being rigid, and does not operate with the presence of water molecules in the binding pocket, hence these methods are unable to identify experimental poses, and proper docking scores for mGlu5 ligands. Validation by retrospective virtual screening campaigns was used to show that the proposed scheme outperforms conventional docking strategies and could be useful for other systems where these effects are important.
Methods
Ligand preparation, Protein preparation, Docking: mGlu5 receptor negative allosteric modulators were downloaded from the ChEMBL database [36]. Compounds with lower than 1 µM IC50 or EC50 values on the human mGlu5 receptor were selected. The highest activity was considered for compounds with multiple reported activities. An active set with 309 compounds was compiled in this way. These ligands were clustered using Canvas [37, 38], based on fingerprint similarity using Daylight invariant atom types; bonds were distinguished by bond order and cyclic aliphatic bonds were distinguished from acyclic aliphatic bonds. Twelve clusters were obtained this way, out of which six were used in the subsequent study; three clusters with acetylenic linker containing 91 compounds (Table S1), two clusters with fenobam analogues containing 63 compounds (Table S2) and one cluster with HTL-14242 analogues containing 36 compounds (Table S3). Other clusters of actives were not used as they were structurally different from the compounds whose complex structures were used for docking. 50 decoy molecules were generated for each active ligand using DUD-E [31, 32]. The active ligands and decoys were prepared for docking with the default options of Ligprep (Schrödinger Suite, 2018-1 [39] and using Maestro (version: 11.5.011) [40]). The proteins were prepared using default settings of Schrödinger Protein Preparation Wizard [39, 41], that includes the optimization of the hydrogen bond network.
Ligands were removed from the complexes and the empty protein cavities were solvated with WaterFLAP using iterative water hotspot identification with molecular interaction field analysis (GRID) [42, 43] and short molecular dynamics simulation (WaterFLAP version 2.2.1). Waters clashing with the ligands of the complex were removed and the remaining water molecules were optimized by water perturbation analysis within WaterFLAP.
The molecular docking was performed with Glide [44] using standard precision and default settings. Glide’s docking score was used to rank compounds and virtual screening results were evaluated by calculating enrichments factors using KNIME [45]. Enrichment factors of the top scored 1% compounds are calculated as EF1% = [TP1%/(TP1% + FP1%)]/[ACTIVETOT/(ACTIVETOT + DECOYTOT)], where TP1% and FP1% are the number of true positives (actives) and the number of false positives (decoys), respectively, in the top 1% of the ranked ligand set, ACTIVETOT and DECOYTOT are the total number of actives and decoys, respectively, in the database.
Results and discussion
Virtual screening was performed using protein structures both without water molecules and with various sets of water molecules. The protein structures were derived from the mGlu5 receptor complexes with mavoglurant (PDB ID: 4OO9, res.: 2.6 Å) [7], HTL-14242 (PDB ID: 5CGD, res.: 2.603 Å) [8] and fenobam (PDB ID: 6FFH, res.: 2.65 Å) [9]. Water molecules were generated by WaterFLAP. It was found that WaterFLAP well reproduces the position of crystallographic waters and, in addition, it predicts the presence of additional water molecules. Some of these waters are close to the ligands and are expected to directly affect ligand binding and to influence virtual screening. Waters used in each complex are shown in Fig. 3. Water HOH 1 generated by WaterFLAP is close to the water molecule that is consistently present in all available X-ray structures (HOH 4126 in 4OO9 crystal structure; see Fig. 3). It is worth noting that WaterFLAP predicts that the free-energy of this water decreases upon ligand binding.
mGlu5 receptor binding pocket with mavoglurant (cyan), HTL-14242 (orange) and fenobam (light green). Water molecules generated by WaterFLAP (HOH1-6) and used in the docking studies are shown as red balls and X-ray water (HOH 4126) is shown as purple ball. Trp7856.50c that adopts different conformation in the mavoglurant complex (shown as green sticks) as compared to the HTL-14242 and fenobam complexes (shown as yellow sticks) is also shown
We identified five water molecules (waters 1, 2, 3, 5 and 6) in the mavoglurant-mGlu5 receptor complex within 4 Å separation from the ligand out of which four were common in all three crystal structures (waters 1–3, and 6). Water 5 is shared with the fenobam, but not with the HTL-14242 complex since in the latter it is displaced by the pyrimidine ring that occupies a position not filled in the mavoglurant and fenobam complexes. Water 4 is present in the fenobam and HTL-14242 complexes, but it is not found in the mavoglurant complex since it is replaced by the carboxylate substituent. Thus, five water sites are identified in the mavoglurant and HTL-14242, and six water sites are identified in the fenobam complex derived protein.
Virtual screening studies were performed with the protein structures obtained from the three complexes after the removal of the ligands and all crystallographic water molecules. In addition to these structures, further structures were created by adding water molecules generated by WaterFLAP around the ligands in various combinations. In the first step one water molecule was added to the empty structures. Five structures with one water molecules were obtained for the mavoglurant and the HTL-14242, and six structures were obtained for the fenobam complex derived protein structure. In the next step, the three single water containing structures, each derived from different complexes and best performing in docking studies were selected and a further water molecule was added in each possible position shown in Fig. 3. Structures with three water molecules were prepared in the same way. This procedure resulted in one empty and fourteen water containing structures from the mavoglurant complex derived protein. In this case, structures with water pairs 6-1 and 6-5 performed similarly (see below) and both were used to generate structures with three water molecules. Fifteen water containing structures were generated for fenobam where six water positions were identified. Finally, five water containing structures were generated for HTL-14242. Owing to the good performance of the single water containing structures no further water molecules were added. Altogether, 37 virtual screenings were carried out using 3 structures without water and 34 structures with various number of water molecules.
Virtual screening with mavoglurant–mGlu5 receptor complex derived protein structures
Virtual screening was performed for the empty protein and for structures with one, two and three waters. Receiver operating characteristics curves are depicted in Fig. S1, enrichment factors (EF) for the top 1%, 2% and 5% ranked compounds and other characteristic values of the virtual screening are shown in Table 1.
The mavoglurant structures are able to accommodate over 90% of the compounds in the database. The enrichment factor for the empty mavoglurant structure is 8.9 and this is slightly lower than the enrichment factors obtained with most of the water containing structures. The presence of any single water molecule has a small effect on the enrichments observed for the top-scored 1, 2 and 5% compounds. Water at position 6 was selected for combining it with other water molecules. No significant improvement in the enrichment factors were observed either with two or three water molecules. The structures with two water molecules in the 6-1 and 6-5 positions performed similarly, and both were used in generating structures with three water molecules. Considering EF1%, EF2% and EF5% values, water combination 6-1-3 was found to be the best, slightly outperforming other mavoglurant complex derived structures.
Virtual screening with fenobam–mGlu5 receptor complex derived protein structures
The fenobam complex derived protein structure contains the Trp7856.50 sidechain pointing inwards the helical bundle. Therefore, these structures have smaller allosteric site than do mavoglurant derived structures. Receiver operating characteristics curves are depicted in Fig. S2, enrichment factors for the top 1%, 2% and 5% ranked compounds and other characteristic values of the virtual screening are shown in Table 2.
The protein is able to accommodate around 50% of the compounds and this is in contrast to the 90% obtained with the mavoglurant complex derived structures. Although both actives and decoys are similar to fenobam [32], the shape and size of the allosteric site and, in particular, the position of Trp7856.50 that points inward, limits the number of successfully docked compounds. Docking into the empty protein could not identify any active among the top ranked 1%, however, the presence of water 1 and 6, and especially water 5 improves the performance. The combination of water 5 with other water molecules, except with water 4, gives double digit enrichment. The 5-3 combination gives consistently the highest enrichments at 1, 2 and 5% ranked compounds. Inclusion of a third water molecule also yields good enrichment factors, however, they are not superior to the 5-3 pair, except the 5-3-6 combination at 2% ranked compounds with an enrichment factor of 12.
Virtual screening with HTL-14242–mGlu5 receptor complex derived protein structures
Results obtained for structures derived from the HTL-14242 complex are presented in Table 3 and in Fig. S3.
The total number of successfully docked compounds is around 80% of the total database irrespective of the number of water molecules included. This is smaller than it was seen for mavoglurant and somewhat higher than it was observed for fenobam. The differences can be partly attributed to the different conformation of Trp7856.50 in the mavoglurant compared to the fenobam and HTL-14242 complexes. The latter structures share “swung-in” Trp7856.50 conformation that yields a significant reduction of the pocket size. The conformational change of this conserved Trp residue contributes to the activation of several class A GPCRs [47, 48] while both conformations appear to be accessible in mGlu5 PAM complexes [29, 30], and the conformation is ligand dependent in mGlu5 NAM complexes [7,8,9]. However, Trp7856.50 is not the only determinant of the size and the shape of the allosteric pocket. Fenobam and HTL-14242 pockets differ in shape (Fig. 4) owing to the different orientation of Ser6543.39 and Val8067.36 side chains. Besides the large number of successfully docked molecules, a high enrichment of 51 in the top 1% of the ranked compounds was achieved with the empty HTL-14242 structure. This is the maximum enrichment that can be obtained at 1%, as all compounds in the first 1% are active. Some improvements in the EF2% and EF5% values were observed with the addition of a water molecule. The best early enrichments are achieved with water 4, with a maximal available EF1%, the highest EF2% and a near to highest EF5%. Since the docking results both without water and with a single water molecule are close to the optimum, no further water combinations were tested. It has to be noted, that the very high enrichments in this case can be attributed to the high similarity of the active compounds that all belong to the series investigated in a structure–activity relationship study around HTL-14242. However, the successful docking of 80% of the compounds shows that the outstanding enrichment is not the consequence of having decoys not accommodated in the allosteric pocket, rather it is the consequence of the efficient distinction between active and inactive compounds by the docking-scoring procedure.
The separation of ligands and using an appropriate protein structure with optimally selected water molecules outperforms the conventional approach. Virtual screening campaigns with all ligands using any of the three protein structures give vastly inferior early enrichments (Table S4). Ligands assigned to each protein structure may belong to several clusters as it is the case for mavoglurant (3 clusters) and fenobam analogues (2 clusters). The analysis of virtual screening performances separately for each cluster (Tables S5–7) provides indications on how much similarity to the X-ray ligands is needed for a successful virtual screening. As an example, actives in cluster 12 are more similar to mavoglurant than those of cluster 11 (median similarity 0.532 vs. 0.078), and enrichment factors of cluster 12 are superior to those of cluster 11. This suggests that similarity information is useful in guiding prospective virtual screening applications.
A detailed analysis of the water molecules yielding improved VS performance revealed that they are ligand dependent, and their common feature is the presence of hydrogen bonds to the ligands. Mavoglurant forms no hydrogen bond to any of the water molecules, and a modest increase of enrichment factors was achieved compared to the empty protein structure. HTL-14242 gave high enrichment factors already with the empty protein, and the EF2% enrichment was further improved in the presence of HOH 4, a water molecule hydrogen bonded to HTL-14242 and also to Ile6513.36 and Asn7475.47. The highest improvement of enrichments for fenobam was observed with the addition of HOH 5 (EF1% increased from 0 to 12.8), and this was further increased when both HOH 5 and HOH 3 were added. HOH 5 is hydrogen bonded to fenobam and to Ile6513.36, and HOH 3 forms hydrogen bonds to fenobam and to both Ser8057.36 and HOH 2. Other water molecules having smaller effect on enrichments are not hydrogen-bonded to fenobam. The HOH 1 water molecule consistently observed in all mGlu5 receptor X-ray structures does not form hydrogen-bond with any of the ligands and its presence was not found to improve virtual screening performance. Although this water molecule appears to contribute to the structural stability of the receptor and even more to its complexes as suggested by WaterFLAP calculations, it does not have direct interactions with the ligands investigated and its presence does affect virtual screening performance. The observations suggest a guideline for selecting water molecules in the virtual screening of the proposed protocol; waters hydrogen-bonded to the ligand are most likely to beneficially affect enrichment factors, and the number and position of these water molecules are ligand dependent.
Based on the above results a virtual screening protocol is proposed that has two key elements to assure that both protein flexibility and the role of water molecules are well accounted for. It includes the separation of the ligands into compound series and docking each series into a distinct protein structure having appropriate conformation to accommodate the particular series of compounds. Restricting compounds to those structurally related to a bound ligand of the protein conformation used for docking decreases false negative rates, while using a single protein structure for a particular compound series decreases false positive rates. The use of an experimental protein structure complexed with a ligand structurally close to the compounds to be docked is reminiscent to self-docking, where a ligand is docked into the experimental protein structure of its complex. In contrast, in cross-docking, a compound is docked into a protein structure extracted from the complex with another ligand. The better performance of self-docking over cross-docking is well-established [49, 50] and it is more pronounced for scoring and ranking than for docking accuracy [51]. Protein flexibility was shown to increase the difficulty of selecting the appropriate protein structure for successful docking [52, 53], and the excessive sensitivity of the mGlu5 receptor binding pocket on the bound ligand calls for the judicious selection of mGlu5 target structures. The other key element of the procedure is to place computationally generated water molecules into the binding pocket. The evaluation of retrospective virtual screenings performed with various combinations of water molecules informs us on the optimal arrangement of water molecules. The proposed protocol is depicted in Fig. 5.
It was demonstrated above that separating ligands into series and docking them into distinct protein structures with selected water molecules yields good early enrichments. The separation of ligands into structurally similar series is motivated by the induced fit binding to the mGlu5 receptor observed in X-ray structures. Although induced fit binding to proteins is frequently observed, mGlu5 receptor appears to be extremely sensitive to small structural variations of the ligand as it is exemplified by the binding of mavoglurant and M-MPEP. These two compounds are close analogues (Fig. 1), nevertheless, their cross-docking (the docking of M-MPEP into the protein structure that binds mavoglurant and vice versa) fails to straightforwardly reproduce the experimental complexes. Another feature of mGlu5 receptor is the presence of a functional water network in the allosteric site. Although the effect of binding site water molecules on ligand binding is generally observed, this appears to be enhanced in the allosteric site of mGlu5 receptor owing to its narrow shape and well structured, functionally relevant water network. Therefore, the presence and the interactions of the water molecules have profound effect on ligand binding, and this has to be taken into account in the structure based virtual screening of the mGlu5 receptor.
The effect of protein flexibility and the interaction with water molecules on ligand binding is a general feature of GPCR allosteric sites. Experimental structural data obtained with and without bound allosteric ligands show that these sites are sensitive to protein conformational changes and to interactions with water molecules. Therefore, the proposed protocol that is able to account for these effects is expected to find use in the virtual screening of a wide range of GPCR allosteric sites.
Conclusion
Our analysis focused on the virtual screening performance of mGlu5 receptor protein structures derived from complexes with negative allosteric modulators having varying scaffolds. Important factors in ligand binding to mGlu5 receptor is protein flexibility and interactions with the water network in the allosteric pocket. Protein flexibility results in induced fit binding and this necessitates the separation of ligands into structurally similar series and docking each series to a protein conformation derived from a complex with a member of the series. We considered three ligands with available mGlu5 receptor X-ray structures. One of the compounds, mavoglurant, contains an acetylenic linker and another, fenobam, contains a urea linker both render the ligands linear. The third ligand, HTL-14242, has a heterocyclic linker that results in a bent shape. Docking compound series into the corresponding protein conformation gave varying enrichments. We found that the inclusion of water molecules in the docking protocol is able to improve the docking performance for all structures studied. This site is a functional water channel and the interaction of the waters with the protein, with the ligand and with each other contribute importantly to the ligand binding. Therefore, the inclusion of waters in the protein structure is beneficial for the performance of the virtual screening.
The experience gained with the retrospective hit identification by virtual screening can be translated to a virtual screening protocol to identify allosteric ligands for the mGlu5 receptor. The principal elements of the protocol are the use of several protein structures derived from complexes of ligands with distinct shape or chemotype and to identify water positions by optimizing the performance of small scale retrospective virtual screening calculations. The protocol allows the extension of available set of allosteric ligands and to find novel chemical starting points for mGlu5 receptor. The protocol takes into account protein flexibility and the presence of water molecules in the binding site and is expected to be useful for other systems where these effects are important.
References
Lionta E, Spyrou G, Vassilatis DK, Cournia Z (2014) Structure-based virtual screening for drug discovery: principles, applications and recent advances. Curr Top Med Chem 14:1923–1938. https://doi.org/10.2174/1568026614666140929124445
Kooistra AJ, Roumen L, Leurs R et al (2013) From heptahelical bundle to hits from the haystack: structure-based virtual screening for GPCR ligands. Methods Enzymol 522:279–336. https://doi.org/10.1016/B978-0-12-407865-9.00015-7
Bartuzi D, Kaczor AA, Matosiuk D (2017) Signaling within allosteric machines: signal transmission pathways inside G protein-coupled receptors. Molecules. https://doi.org/10.3390/molecules22071188
Thal DM, Glukhova A, Sexton PM, Christopoulos A (2018) Structural insights into G-protein-coupled receptor allostery. Nature 559:45–53. https://doi.org/10.1038/s41586-018-0259-z
Hauser AS, Attwood MM, Rask-Andersen M et al (2017) Trends in GPCR drug discovery: new agents, targets and indications. Nat Rev Drug Discov 16:829–842. https://doi.org/10.1038/nrd.2017.178
Wakefield AE, Mason JS, Vajda S, Keserű GM (2019) Analysis of tractable allosteric sites in G protein-coupled receptors. Sci Rep 9:6180. https://doi.org/10.1038/s41598-019-42618-8
Doré AS, Okrasa K, Patel JC et al (2014) Structure of class C GPCR metabotropic glutamate receptor 5 transmembrane domain. Nature 511:557–562. https://doi.org/10.1038/nature13396
Christopher JA, Aves SJ, Bennett KA et al (2015) Fragment and structure-based drug discovery for a class C GPCR: discovery of the mGlu5 negative allosteric modulator HTL14242 (3-Chloro-5-[6-(5-fluoropyridin-2-yl)pyrimidin-4-yl]benzonitrile). J Med Chem 58:6653–6664. https://doi.org/10.1021/acs.jmedchem.5b00892
Christopher JA, Orgován Z, Congreve M et al (2018) Structure-based optimization strategies for G protein-coupled receptor (GPCR) allosteric modulators: a case study from analyses of new metabotropic glutamate receptor 5 (mGlu 5) X-ray structures. J Med Chem. https://doi.org/10.1021/acs.jmedchem.7b01722
Koehl A, Hu H, Feng D et al (2019) Structural insights into the activation of metabotropic glutamate receptors. Nature 566:79–84. https://doi.org/10.1038/s41586-019-0881-4
Alagarsamy S, Sorensen SD, Conn PJ (2001) Coordinate regulation of metabotropic glutamate receptors. Curr Opin Neurobiol 11:357–362
Niswender CM, Conn PJ (2010) Metabotropic glutamate receptors: physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol 50:295–322. https://doi.org/10.1146/annurev.pharmtox.011008.145533
Congreve M, Oswald C, Marshall FH (2017) Applying structure-based drug design approaches to allosteric modulators of GPCRs. Trends Pharmacol Sci 38:837–847. https://doi.org/10.1016/j.tips.2017.05.010
Lindsley CW, Emmitte KA, Hopkins CR et al (2016) Practical strategies and concepts in GPCR allosteric modulator discovery: recent advances with metabotropic glutamate receptors. Chem Rev 116:6707–6741. https://doi.org/10.1021/acs.chemrev.5b00656
Llinas del Torrent C, Casajuana-Martin N, Pardo L et al (2019) Mechanisms underlying allosteric molecular switches of metabotropic glutamate receptor 5. J Chem Inf Model 59:2456–2466. https://doi.org/10.1021/acs.jcim.8b00924
Vranesic I, Ofner S, Flor PJ et al (2014) AFQ056/mavoglurant, a novel clinically effective mGluR5 antagonist: identification, SAR and pharmacological characterization. Bioorg Med Chem 22:5790–5803. https://doi.org/10.1016/j.bmc.2014.09.033
Gasparini F, Lingenhöhl K, Stoehr N et al (1999) 2-Methyl-6-(phenylethynyl)-pyridine (MPEP), a potent, selective and systemically active mGlu5 receptor antagonist. Neuropharmacology 38:1493–1503. https://doi.org/10.1016/S0028-3908(99)00082-9
Wållberg A, Nilsson K, Österlund K et al (2006) Phenyl ureas of creatinine as mGluR5 antagonists. A structure–activity relationship study of fenobam analogues. Bioorg Med Chem Lett 16:1142–1145. https://doi.org/10.1016/j.bmcl.2005.11.092
Pagano A, Ruegg D, Litschig S et al (2000) The non-competitive antagonists 2-methyl-6-(phenylethynyl)pyridine and 7-hydroxyiminocyclopropan[b]chromen-1a-carboxylic acid ethyl ester interact with overlapping binding pockets in the transmembrane region of group I metabotropic glutamate receptors. J Biol Chem 275:33750–33758. https://doi.org/10.1074/jbc.M006230200
Kaae BH, Harpsøe K, Kvist T et al (2012) Structure–activity relationships for negative allosteric mGluR5 modulators. ChemMedChem 7:440–451. https://doi.org/10.1002/cmdc.201100578
Dalton JAR, Gómez-Santacana X, Llebaria A, Giraldo J (2014) Computational analysis of negative and positive allosteric modulator binding and function in metabotropic glutamate receptor 5 (in)activation. J Chem Inf Model 54:1476–1487. https://doi.org/10.1021/ci500127c
Anighoro A, Graziani D, Bettinelli I et al (2015) Insights into the interaction of negative allosteric modulators with the metabotropic glutamate receptor 5: discovery and computational modeling of a new series of ligands with nanomolar affinity. Bioorg Med Chem 23:3040–3058. https://doi.org/10.1016/j.bmc.2015.05.008
Harpsøe K, Isberg V, Tehan BG et al (2015) Selective negative allosteric modulation of metabotropic glutamate receptors—a structural perspective of ligands and mutants. Sci Rep 5:13869. https://doi.org/10.1038/srep13869
Vijaya Prabhu S, Singh SK (2019) E-pharmacophore-based screening of mGluR5 negative allosteric modulators for central nervous system disorder. Comput Biol Chem 78:414–423. https://doi.org/10.1016/J.COMPBIOLCHEM.2018.12.016
Llinas Del Torrent C, Pérez-Benito L, Tresadern G (2019) Computational drug design applied to the study of metabotropic glutamate receptors. Molecules. https://doi.org/10.3390/molecules24061098
Gatica EA, Cavasotto CN (2012) Ligand and decoy sets for docking to G protein-coupled receptors. J Chem Inf Model 52:1–6. https://doi.org/10.1021/ci200412p
Ivanov AA, Barak D, Jacobson KA (2009) Evaluation of homology modeling of G-protein-coupled receptors in light of the A2A adenosine receptor crystallographic structure. J Med Chem 52:3284–3292. https://doi.org/10.1021/jm801533x
Lenselink EB, Beuming T, Sherman W et al (2014) Selecting an optimal number of binding site waters to improve virtual screening enrichments against the adenosine A2A receptor. J Chem Inf Model 54:1737–1746. https://doi.org/10.1021/ci5000455
Jójárt B, Orgován Z, Márki Á et al (2019) Allosteric activation of metabotropic glutamate RECEPTOR 5. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2019.1638302
Cong X, Chéron J-B, Golebiowski J et al (2019) Allosteric modulation mechanism of the mGluR5 transmembrane domain. J Chem Inf Model. https://doi.org/10.1021/acs.jcim.9b00045
Huang N, Shoichet KB, Irwin JJ (2006) Benchmarking sets for molecular docking. J Med Chem. https://doi.org/10.1021/JM0608356
Mysinger MM, Carchia M, Irwin JJ, Shoichet BK (2012) Directory of useful decoys, enhanced (DUD-E): better ligands and decoys for better benchmarking. J Med Chem 55:6582–6594. https://doi.org/10.1021/jm300687e
Feng Z, Ma S, Hu G, Xie X-Q (2015) Allosteric binding site and activation mechanism of class C G-protein coupled receptors: metabotropic glutamate receptor family. AAPS J 17:737–753. https://doi.org/10.1208/s12248-015-9742-8
Mølck C, Harpsøe K, Gloriam DE et al (2014) mGluR5: exploration of orthosteric and allosteric ligand binding pockets and their applications to drug discovery. Neurochem Res 39:1862–1875
Molck C, Harpsoe K, Gloriam DE et al (2012) Pharmacological characterization and modeling of the binding sites of novel 1,3-bis(pyridinylethynyl)benzenes as metabotropic glutamate receptor 5-selective negative allosteric modulators. Mol Pharmacol 82:929–937. https://doi.org/10.1124/mol.112.078808
Gaulton A, Hersey A, Nowotka M et al (2017) The ChEMBL database in 2017. Nucleic Acids Res 45:D945–D954. https://doi.org/10.1093/nar/gkw1074
Duan J, Dixon SL, Lowrie JF, Sherman W (2010) Analysis and comparison of 2D fingerprints: insights into database screening performance using eight fingerprint methods. J Mol Graph Model 29:157–170. https://doi.org/10.1016/J.JMGM.2010.05.008
Sastry M, Lowrie JF, Dixon SL, Sherman W (2010) Large-scale systematic analysis of 2D fingerprint methods and parameters to improve virtual screening enrichments. J Chem Inf Model 50:771–784. https://doi.org/10.1021/ci100062n
Schrödinger Suite 2018-1 (2016) Protein preparation wizard. Epik, Schrödinger, LLC, New York
Maestro (2018) Schrödinger, LLC, New York, NY
Sastry GM, Adzhigirey M, Day T et al (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/s10822-013-9644-8
Goodford PJ (1985) A computational procedure for determining energetically favorable binding sites on biologically important macromolecules. J Med Chem 28:849–857. https://doi.org/10.1021/jm00145a002
Sciabola S, Stanton RV, Mills JE et al (2010) High-throughput virtual screening of proteins using GRID molecular interaction fields. J Chem Inf Model 50:155–169. https://doi.org/10.1021/ci9003317
Glide (2017) Schrödinger, LLC, New York
Berthold MR, Cebron N, Dill F et al (2008) KNIME: the Konstanz information miner. Springer, Berlin, pp 319–326
Nicholls A (2014) Confidence limits, error bars and method comparison in molecular modeling. Part 1: the calculation of confidence intervals. J Comput Aided Mol Des 28:887–918. https://doi.org/10.1007/s10822-014-9753-z
Trzaskowski B, Latek D, Yuan S et al (2012) Action of molecular switches in GPCRs–theoretical and experimental studies. Curr Med Chem 19:1090–1109. https://doi.org/10.2174/092986712799320556
Ahuja S, Smith SO (2009) Multiple switches in G protein-coupled receptor activation. Trends Pharmacol Sci 30:494–502. https://doi.org/10.1016/j.tips.2009.06.003
Ramírez D, Caballero J (2018) Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules 23:1038. https://doi.org/10.3390/molecules23051038
Erickson JA, Jalaie M, Robertson DH et al (2004) Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J Med Chem 47:45–55. https://doi.org/10.1021/jm030209y
Cavasotto CN, Abagyan RA (2004) Protein flexibility in ligand docking and virtual screening to protein kinases. J Mol Biol 337:209–225. https://doi.org/10.1016/J.JMB.2004.01.003
Gathiaka S, Liu S, Chiu M et al (2016) D3R grand challenge 2015: evaluation of protein–ligand pose and affinity predictions. J Comput Aided Mol Des 30:651–668. https://doi.org/10.1007/s10822-016-9946-8
Bajda M, Jończyk J, Malawska B, Filipek S (2014) Application of computational methods for the design of BACE-1 inhibitors: validation of in silico modelling. Int J Mol Sci 15:5128–5139. https://doi.org/10.3390/ijms15035128
Acknowledgements
Open access funding provided by MTA Research Centre for Natural Sciences (MTA TTK). This work has been supported by the National Brain Research Program (2017-1.2.1-NKP-2017-00002) of the National Research, Development and Innovation Office, Hungary.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Orgován, Z., Ferenczy, G.G. & Keserű, G.M. The role of water and protein flexibility in the structure-based virtual screening of allosteric GPCR modulators: an mGlu5 receptor case study. J Comput Aided Mol Des 33, 787–797 (2019). https://doi.org/10.1007/s10822-019-00224-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-019-00224-w