Abstract
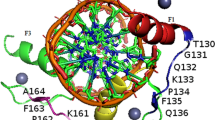
Molecular dynamics and MM_GBSA energy calculations on various zinc finger proteins containing three and four fingers bound to their target DNA gave insights into the role of each finger in the DNA binding process as part of the protein structure. The wild type Zif 268 (PDB code: 1AAY) gave a ΔG value of − 76.1 (14) kcal/mol. Zinc fingers ZF1, ZF2 and ZF3 were mutated in one experiment and in another experiment one finger was cut and the rest of the protein was studied for binding. The ΔΔG values for the Zinc Finger protein with both ZF1 and ZF2 mutated was + 80 kcal/mol, while mutating only ZF1 the ΔΔG value was + 52 kcal/mol (relative to the wild type). Cutting ZF3 and studying the protein consisting only of ZF1 linked to ZF2 gave a ΔΔG value of + 68 kcal/mol. Upon cutting ZF1, the resulting ZF2 linked to ZF3 protein gave a ΔΔG value of + 41 kcal/mol. The above results shed light on the importance of each finger in the binding process, especially the role of ZF1 as the anchoring finger followed in importance by ZF2 and ZF3. The energy difference between the binding of the wild type protein Zif268 (1AAY) and that for individual finger binding to DNA according to the formula: ΔΔGlinkers, otherstructuralfactors = ΔGzif268 − (ΔGF1+F2+F3) gave a value = − 44.5 kcal/mol. This stabilization can be attributed to the contribution of linkers and other structural factors in the intact protein in the DNA binding process. DNA binding energies of variant proteins of the wild type Zif268 which differ in their ZF1 amino acid sequence gave evidence of a good relationship between binding energy and recognition and specificity, this finding confirms the reported vital role of ZF1 in the ZF protein scanning and anchoring to the target DNA sequence. The role of hydrogen bonds in both specific and nonspecific amino acid-DNA contacts is discussed in relation to mutations. The binding energies of variant Zinc Finger proteins confirmed the role of ZF1 in the recognition, specificity and anchoring of the zinc finger protein to DNA.










Similar content being viewed by others
References
Wu H, Yang WP, Barbas CF (1995) Building zinc fingers by selection: toward a therapeutic application. Proc Natl Acad Sci USA 92:344–348. https://doi.org/10.1073/pnas.92.2.344
Elrod-Erickson M, Benson TE, Pabo CO (1998) High-resolution structures of variant Zif268–DNA complexes: implications for understanding zinc finger–DNA recognition. Structure 6:451–464
Lee J, Kim J-S, Seok C (2010) Cooperativity and specificity of Cys2His2 zinc finger protein-DNA interactions: a molecular dynamics simulation study. J Phys Chem B 114:7662–7671. https://doi.org/10.1021/jp1017289
Cheng X, Boyer JL, Juliano RL (1997) Selection of peptides that functionally replace a zinc finger in the Sp1 transcription factor by using a yeast combinatorial library. Proc Natl Acad Sci USA 94:14120–14125
Dreier B, Fuller RP, Segal DJ et al (2005) Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem 280:35588–35597. https://doi.org/10.1074/jbc.M506654200
Greisman H (1997) A general strategy for selecting high-affinity zinc finger proteins for diverse DNA target sites. Science 275:657–661. https://doi.org/10.1126/science.275.5300.657
Hamed MY, Arya G (2016) Zinc finger protein binding to DNA: an energy perspective using molecular dynamics simulation and free energy calculations on mutants of both zinc finger domains and their specific DNA bases. J Biomol Struct Dyn 34:919–934
Höglund A, Kohlbacher O (2004) From sequence to structure and back again: approaches for predicting protein-DNA binding. Proteome Sci 2:3. https://doi.org/10.1186/1477-5956-2-3
Elrod-Erickson M, Rould M, Nekludova L, Pabo CO (1996) Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure 4:1171–1180
Fu F, Sander JD, Maeder M et al (2009) Zinc finger database (ZiFDB): a repository for information on C2H2 zinc fingers and engineered zinc-finger arrays. Nucleic Acids Res. https://doi.org/10.1093/nar/gkn606
Papworth M, Kolasinska P, Minczuk M (2006) Designer zinc-finger proteins and their applications. Gene 366:27–38
Hamed SN, Sanie M, Frank WS et al (2015) C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol 33:555–562. https://doi.org/10.1038/nbt.3128
Pabo CO, Peisach E, Grant RA (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. https://doi.org/10.1146/annurev.biochem.70.1.313
Zandarashvili L, Vuzman D, Esadze A. et al (2012) PNAS Plus: asymmetrical roles of zinc fingers in dynamic DNA-scanning process by the inducible transcription factor Egr-1. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1121500109
Chen C, Esadze A, Zandarashvili L et al (2015) Dynamic equilibria of short-range electrostatic interactions at molecular interfaces of protein-DNA complexes. J Phys Chem Lett 6:2733–2737. https://doi.org/10.1021/acs.jpclett.5b01134
Pearlman DA, Case DA, Caldwell JW, Ross WR, Chealtham TE, DeBolt S, Ferguson D., Seibel G, Kollman P (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculation to simulate the structural and energetic properties of molecules. Comput Phys Commun 91(1–3):1–41
Case DA, Pearlman JW, Caldwell TE, Cheatham J III, Wang WSR, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vencent JJ, Crawley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pietera J, Massova I, Singh GLS, Weiner PK (2006) Amber 9, University of California, San Francisco
DeLano WL (2002) The PyMOL molecular graphics system. Delano Scientific, San Carlos
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Lee J, Kim J-S, Seok C (2010) Cooperativity and specificity of Cys2His2 zinc finger protein-DNA interactions: a molecular dynamics simulation study. J Phys Chem B 114:7662–7671
Wolfe S, Nekludova L, Pabo CO (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29:183–212. https://doi.org/10.1146/annurev.biophys.29.1.183
Segal DJ, Crotty JW, Bhakta MS et al (2006) Structure of aart, a designed six-finger zinc finger peptide, bound to DNA. J Mol Biol 363:405–421. https://doi.org/10.1016/j.jmb.2006.08.016
Nguyen-Hackley DH, Ramm E, Taylor CM, Joung JK, Dervan PB, Pabo CO (2004) Allosteric inhibition of zinc-finger binding in the major groove of DNA by minor-groove binding ligands. Biochemistry 43:3880–3890
Hamed MY, Arya G (2015) Zinc finger protein binding to DNA: an energy perspective using molecular dynamics simulation and free energy calculations on mutants of both zinc finger domains and their specific DNA bases. J Biomol Struct Dyn. https://doi.org/10.1080/07391102.2015.1068224
Iuchi S (2005) C2H2 zinc fingers as DNA binding domains. In: Zinc finger proteins. Springer, New York, pp 7–13
Ryan RF, Darby MK (1998) The role of zinc finger linkers in p43 and TFIIIA binding to 5S rRNA and DNA. Nucleic Acids Res 26:703–709. https://doi.org/10.1093/Nar/26.3.703 doi
Peisach E, Pabo CO (2003) Constraints for zinc finger linker design as inferred from X-ray crystal structure of tandem Zif268–DNA complexes. J Mol Biol 330:1–7. https://doi.org/10.1016/S0022-2836(03)00572-2
Laity JH, Dyson HJ, Wright PE (2000) DNA-induced α-helix capping in conserved linker sequences is a determinant of binding affinity in Cys 2-His 2 zinc fingers. J Mol Biol 295:719–727
Nagaoka M, Nomura W, Shiraishi Y, Sugiura Y (2001) Significant effect of linker sequence on DNA recognition by multi-zinc finger protein. Biochem Biophys Res Commun 282:1001–1007
Anand P, Schug A, Wenzel W (2013) Structure based design of protein linkers for zinc finger nuclease. FEBS Lett 587:3231–3235. https://doi.org/10.1016/j.febslet.2013.08.015
Liu J, Stormo GD (2008) Context-dependent DNA recognition code for C2H2 zinc-finger transcription factors. Bioinformatics 24:1850–1857. https://doi.org/10.1093/bioinformatics/btn331
Pabo CO, Nekludova L (2000) Geometric analysis and comparison of protein-DNA interfaces: why is there no simple code for recognition? J Mol Biol 301:597–624. https://doi.org/10.1006/jmbi.2000.3918
Selvaraj S, Kono H, Sarai A (2002) Specificity of protein-DNA recognition revealed by structure-based potentials: symmetric/asymmetric and cognate/non-cognate binding. J Mol Biol 322:907–915. https://doi.org/10.1016/S0022-2836(02)00846-X
Bates DL, Chen Y, Kim G et al (2008) Crystal structures of multiple GATA zinc fingers bound to DNA reveal new insights into DNA recognition and self-association by GATA. J Mol Biol 381:1292–1306. https://doi.org/10.1016/j.jmb.2008.06.072
Sheu S-Y, Yang D-Y, Selzle HL, Schlag EW (2003) Energetics of hydrogen bonds in peptides. Proc Natl Acad Sci USA 100:12683–12687. https://doi.org/10.1073/pnas.2133366100
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hamed, M.Y. Role of protein structure and the role of individual fingers in zinc finger protein–DNA recognition: a molecular dynamics simulation study and free energy calculations. J Comput Aided Mol Des 32, 657–669 (2018). https://doi.org/10.1007/s10822-018-0119-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-018-0119-9