Abstract
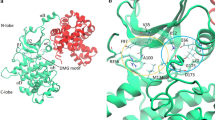
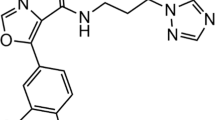
Cdc25 phosphatase B, a potential target for cancer therapy, is inhibited by a series of quinones. The binding site and mode of quinone inhibitors to Cdc25B remains unclear, whereas this information is important for structure-based drug design. We investigated the potential binding site of NSC663284 [DA3003-1 or 6-chloro-7-(2-morpholin-4-yl-ethylamino)-quinoline-5, 8-dione] through docking and molecular dynamics simulations. Of the two main binding sites suggested by docking, the molecular dynamics simulations only support one site for stable binding of the inhibitor. Binding sites in and near the Cdc25B catalytic site that have been suggested previously do not lead to stable binding in 50 ns molecular dynamics (MD) simulations. In contrast, a shallow pocket between the C-terminal helix and the catalytic site provides a favourable binding site that shows high stability. Two similar binding modes featuring protein-inhibitor interactions involving Tyr428, Arg482, Thr547 and Ser549 are identified by clustering analysis of all stable MD trajectories. The relatively flexible C-terminal region of Cdc25B contributes to inhibitor binding. The binding mode of NSC663284, identified through MD simulation, likely prevents the binding of protein substrates to Cdc25B. The present results provide useful information for the design of quinone inhibitors and their mechanism of inhibition.







Similar content being viewed by others
References
Boutros R, Lobjois V, Ducommun B (2007) CDC25 phosphatases in cancer cells: key players? Good targets? Nat Rev Cancer 7:495–507
Munter SD, Köhn M, Bollen M (2013) Challenges and opportunities in the development of protein phosphatase-directed therapeutics. ACS Chem Biol 8:36 – 45
Boutros R, Dozier C, Ducommun B (2006) The when and the wheres of CDC25 phosphatases. Curr Opin Cell Biol 18:185 – 191
Van Vugt MA, Bras A, Medema RH (2004) Polo-like kinase-1 controls recovery from a G2 DNA damage-induced arrest in mammalian cells. Mol Cell 15:799–811
Bugler B, Quaranta M, Aressy B, Brezak MC, Prevost G, Ducommun B (2006) Genotoxic-activated G2-M checkpoint exit is dependent on CDC25B phosphatase expression. Mol Cancer Ther 5:1446–1451
Neely KE, Piwnica-Worms H (2003) Cdc25A regulation: to destroy or not to destroy is that the only question? Cell Cycle 2:455–457
Ferguson AM, White LS, Donovan PJ, Piwnica-Worms H (2005) Normal cell cycle and checkpoint responses in mice and cells lacking cdc25b and cdc25c protein phosphatases. Mol Cell Biol 25:2853–2860
Guo JC, Kleeff J, Li JS, Ding JY, Hammer J, Zhao YP, Giese T, Korc M, Buchler MW, Friess H (2004) Expression and functional significance of CDC25B in human pancreatic ductal adenocarcinoma. Oncogene 23:71–81
Contour-Galcera Marie-Odile, Sidhu A, Prévost G, Bigg D, Ducommun B (2007) What’s new on CDC25 phosphatase inhibitors. Pharmacol Ther 115:1–12
Lavecchia A, Giovanni CD, Novellino E (2010) Inhibitors of Cdc25 phosphatases as anticancer agents: a patent review. Expert Opin Ther Patents 20:405–425
Lavecchia A, Di Giovanni C, Novellino E (2009) Cdc25A and B dual-specifity phosphatase inhibitors: potential agents for cancer therapy. Curr Med Chem 16:1831–1849
Lazo JS, Aslan DC, Southwick EC, Cooley KA, Ducruet AP, Joo B, Vogt A, Wipf P (2001) Discovery and biological evaluation of a new family of potent inhibitors of the dual specificity protein phosphatase cdc25. J Med Chem 44:4042–4049
Guo JX, Parise RA, Joseph E, Lan J, Pan SS, Joo B, Egorin MJ, Wipf P, Lazo JS, Eiseman JL (2007) Pharmacology and antitumor activity of a quinolinedione cdc25 phosphatase inhibitor da3003-1 (NSC 663284). Anticancer Res 27:3067–3074
Lavecchia A, Giovanni CD, Pesapane A, Montuori N, Ragno P, Martucci NM, Masullo M, Vendittis ED, Novellino E (2012) Discovery of new inhibitors of cdc25b dual specificity phosphatases by structure-based virtual screening. J Med Chem 55:4142–4158
Cossy J, Belotti D, Brisson M, Skoko JJ, Wipf P, Lazo JS (2006) Biological evaluation of newly synthesized quinoline-5,8-quinones as Cdc25B inhibitors. Bioorg Med Chem 14:6283 – 6287
Wardman P (1990) Bioreductive activation of quinones: redox properties and thiol reactivity. Free Radic Res Commun 8:219–229
Reynolds RA, Yem AW, Wolfe CL, Deibel MR Jr, Chidester CG, Watenpaugh KD (1999) Crystal Structure of the catalytic subunit of Cdc25B required for G2/M phase transition of the cell cycle. J Mol Biol 293:559–568
Rudolph R (2007) Cdc25 phosphatases: structure, specificity, and mechanism. Biochem 46:3595–3603
Buhrman G, Parker B, Sohn J, Rudolph J, Mattos C (2005) Structural mechanism of oxidative regulation of the phosphatase Cdc25B via an intramolecular disulfide bond. Biochem 44:5307–5316
Jackson MD, Denu JM (2001) Molecular reactions of protein phosphatases: Insights from structure and chemistry. Chem Rev 101:2313–2340
Lavecchia A, Cosconati S, Limongelli V, Novellino E (2006) Modeling of Cdc25B dual specifity protein phosphatase inhibitors: docking of ligands and enzymatic inhibition mechanism. ChemMedChem 1:540–550
Brisson M, Nguyen T, Wipf P, Joo B, Day BW, Skoko JS, Schreiber EM, Foster C, Bansal P, Lazo JS (2005) Redox regulation of Cdc25B by cell-active quinolinediones. Mol Pharmacol 68:1810–1820
Arantes GM (2010) Flexibility and inhibitor binding in Cdc25 phosphatases. Proteins 78:3017–3032
Park H, Carr BI, Li MH, Ham SW (2007) Fluorinated NSC as a Cdc25 inhibitor. Bioorg Med Chem Lett 17:2351–2354
Ko S, Lee W, Lee S, Park H (2008) Nanosecond molecular dynamics simulations of Cdc25B and its complex with a 1,4-naphthoquinone inhibitor: Implications for rational inhibitor design. J Mol Graph Model 27:13–19
Braud E, Goddard ML, Kolb S, Brun MP, Mondésert O, Quaranta M, Gresh N, Ducommun B, Garbay C (2008) Novel naphthoquinone and quinolinedione inhibitors of CDC25 phosphatase activity with antiproliferative properties. Bioorg Med Chem 16:9040–9049
Lengauer T, Rareyt M (1996) Computational methods for biomolecular docking. Curr Opin Struct Biol 6:402–406
Brooijmans N, Kuntz ID (2003) Molecular recognition and docking algorithms. Annu Rev Biophys Biomol Struct 32:335–373
Coupez B, Lewis RA (2006) Docking and scoring—theoretically easy, practically impossible? Curr Med Chem 13:2995–3003
Software SYBYL (2008) Verison 8.1. Tripos Associates Inc., St. Louis
Anandakrishnan R, Aguilar B, Onufriev AV (2012) H++ 3.0: automating pKa prediction and the preparation of biomolecular structures for atomistic molecular modeling and simulation. Nucleic Acids Res 40:537–541
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J Comput Chem 16:2785–2791
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461
Goodsell DS, Morris GM, Olson AJ (1996) Automated docking of flexible ligands: applications of autodock. J Mol Recognition 9:1–5
Vanquelef E, Simon S, Marquant G, Garcia E, Klimerak G, Delepine JC, Cieplak P, Dupradeau FY (2011) R.E.D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucl Acids Res 39:511–517
Bayly CI, Cieplak P, Cornell W, Kollman PA (1993) A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem 97:10269–10280
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Salomon-Ferrer R, Case DA, Walker RC (2013) An overview of the Amber biomolecular simulation package. WIREs Comput Mol Sci 3:198–210
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Goetz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathews DH, Seetin MG, Salomon-Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2012) AMBER 12. University of California, San Francisco
Humphrey W, Dalke A, Schulten K (1996) VMD—visual molecular dynamics. J Mol Graph 14:33–38
Seeber M, Cecchini M, Rao F, Settanni G, Caflisch A (2007) Wordom: a program for efficient analysis of molecular dynamics simulations. Bioinformatics 23:2625–2627
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Arantes GM (2008) The catalytic acid in the dephosphorylation of the Cdc2-pTpY/CycA protein complex by Cdc25B phosphatase. J Phys Chem B 112:15244–15247
Fauman EB, Cogswell J, Lovejoy B, Rocque WJ, Holmes W, Montana VG, Piwnica-Worms H, Rink MJ, Saper MA (1998) Crystal structure of the catalytic domain of the human cell cycle control phosphatase, Cdc25A. Cell 93:617–625
Lund G, Cierpicki T (2014) Solution NMR studies reveal no global flexibility in the catalytic domain of Cdc25B. Proteins 82:2889–2895
Sayegh RSR, Tamaki FK, Marana SR, Salinas RK, Arantes GM (2016) Conformational flexibility of the complete catalytic domain of Cdc25B phosphatases. Proteins 84:1567–1575
Lund G, Dudkin S, Borkin D, Ni W, Grembecka J, Cierpicki T (2014) Inhibition of CDC25B phosphatase through disruption of protein–protein interaction. ACS Chem Biol 10(2):390–394
Pu L, Amoscato AA, Bier ME, Lazo JS (2002) Dual G1 and G2 phase inhibition by a novel, selective Cdc25 inhibitor 7-chloro-6-(2-morpholin-4-ylethylamino)-quinoline-5,8-dione. J Biol Chem 277:46877–46885
Sohn J, Parks JM, Buhrman G, Brown P, Kristjansdottir K, Safi A, Edelsbrunner H, Yan WT, Rudolph J (2005) Experimental validation of the docking orientation of Cdc25 with its Cdk2-CycA protein substrate. Biochem 44:16563–16573
Brisson M1, Nguyen T, Vogt A, Yalowich J, Giorgianni A, Tobi D, Bahar I, Stephenson CR, Wipf P, Lazo JS (2004) Discovery and characterization of novel small molecule inhibitors of human Cdc25B dual specificity phosphatase. Mol Pharmacol 66:824–833
Lund G, Dudkin S, Borkin D, Ni W, Grembecka J, Cierpicki T (2015) Inhibition of CDC25B phosphatase through disruption of protein-protein interaction. ACS Chem Biol 10:390–394
Acknowledgements
The authors would like to thank Dr. C. J. Woods for helpful discussion and advice. YG and YL acknowledge financial support from National Science Found for Distinguished Young Scholars of China (21225313), Program for Changjiang Scholars and Innovative Research Team in University (IRT1030). Major State Special Research Project of China (2016YFA0101200), Major State Basic Research Development Program of China (973 Program, 2015CB553701), National Natural Science Foundation of China (NSFC, 21778050), Recruitment Program for Young Professionals (KJ2070000027). MWvdK and AJM thank BBSRC and EPSRC for support (EP/G007705/1; BB/L018756/1; BB/M026280).
Author information
Authors and Affiliations
Corresponding authors
Additional information
The work was performed at the Centre of Computational Chemistry, School of Chemistry, University of Bristol.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ge, Y., van der Kamp, M., Malaisree, M. et al. Identification of the quinolinedione inhibitor binding site in Cdc25 phosphatase B through docking and molecular dynamics simulations. J Comput Aided Mol Des 31, 995–1007 (2017). https://doi.org/10.1007/s10822-017-0073-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-017-0073-y