Abstract
Purpose
The goal of this study is to determine whether any balanced translocation (BT) had been missed by previous karyotyping in patients with unexplained recurrent pregnancy loss (uRPL).
Methods
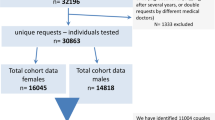
This case series included 48 uRPL-affected couples with normal karyotypes. The embryos from these couples have all undergone preimplantation testing for aneuploidies (PGT-A). Based on the PGT-A’s results, 48 couples could be categorized into two groups: 17 couples whose multiple embryos were detected with similar structural variations (SVs, segmental/complete) and 31 couples without such findings but who did not develop any euploid embryo despite at least three high-quality blastocysts being tested. The peripheral blood sample of each partner was then collected for mate-pair sequencing (MPseq) to determine whether any of them were BT carriers.
Results
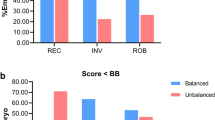
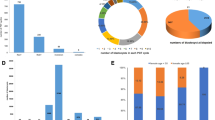
MPseq analyses identified 13 BTs in the 17 couples whose multiple embryos had similar SVs detected (13/17, 76.47%) and three BTs in the 31 couples without euploid embryo obtained (3/31, 9.7%). Among the 16 MPseq-identified BTs, six were missed due to the limited resolution of G-banding karyotyping analysis, and the rest were mostly owing to the similar banding patterns and/or comparable sizes shared by the two segments exchanged.
Conclusion
A normal karyotype does not eliminate the possibility of carrying BT for couples with uRPL. The use of PGT-A allows us to perceive the “carrier couples” missed by karyotyping analysis, providing an increased risk of finding cryptic BTs if similar SVs are always detected on two chromosomes among multiple embryos. Nonetheless, certain carriers with translocated segments of sub-resolution may still go unnoticed.

Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Morin SJ, Eccles J, Iturriaga A, Zimmerman RS. Translocations, inversions and other chromosome rearrangements. Fertil Steril. 2017;107:19–26.
Gupta N, Dalvi R, Koppaka N, Mandava S. Balanced reciprocal translocation: multiple chromosome rearrangements in an infertile female. J Hum Reprod Sci. 2019;12:72–4.
Practice Committee of the American Society for Reproductive Medicine. Evaluation and treatment of recurrent pregnancy loss: a committee opinion. Fertil Steril. 2012;98:1103–11.
Benet J, Oliver-Bonet M, Cifuentes P, Templado C, Navarro J. Segregation of chromosomes in sperm of reciprocal translocation carriers: a review. Cytogenet Genome Res. 2005;111:281–90.
Escudero T, Lee M, Sandalinas M, Munné S. Female gamete segregation in two carriers of translocations involving 2q and 14q. Prenat Diagn. 2000;20:235–7.
Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet. 2012;13:36–46.
Lee H, Gurtowski J, Yoo S, Nattestad M, Marcus S, Goodwin S, et al. Third-generation sequencing and the future of genomics. bioRxiv. 2016 [cited 2022 Jul 23]. p. 048603. Available from: https://www.biorxiv.org/content/10.1101/048603v1.
Smadbeck J, Peterson JF, Pearce KE, Pitel BA, Figueroa AL, Timm M, et al. Mate pair sequencing outperforms fluorescence in situ hybridization in the genomic characterization of multiple myeloma. Blood Cancer J. 2019;9:1–18.
Aypar U, Smoley SA, Pitel BA, Pearce KE, Zenka RM, Vasmatzis G, et al. Mate pair sequencing improves detection of genomic abnormalities in acute myeloid leukemia. Eur J Haematol. 2019;102:87–96.
Dong Z, Yan J, Xu F, Yuan J, Jiang H, Wang H, et al. Genome sequencing explores complexity of chromosomal abnormalities in recurrent miscarriage. Am J Hum Genet. 2019;105:1102–11.
Caspersson T, Zech L, Johansson C, Modest EJ. Identification of human chromosomes by DNA-binding fluorescent agents. Chromosoma. 1970;30:215–27.
Drets ME, Shaw MW. Specific banding patterns of human chromosomes. Proc Natl Acad Sci U S A. 1971;68:2073–7.
Pardue ML, Gall JG. Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci U S A. 1969;64:600–4.
Kawashima E, Farinelli L, Mayer P. Method of nucleic acid amplification. 1998 [cited 2022 Jun 4]. Available from: https://patents.google.com/patent/WO1998044151A1/en.
Kris A. W. DNA sequencing costs: data from the NHGRI Genome Sequencing Program (GSP). DNA Sequencing Costs: Data. [cited 2022 Jun 4]. Available from: https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data.
Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods. 2010;7:461–5.
Schwartz DC, Li X, Hernandez LI, Ramnarain SP, Huff EJ, Wang YK. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–4.
Sundheimer LW, Liu L, Buyalos RP, Hubert G, Al-Safi Z, Shamonki M. Diagnosis of parental balanced reciprocal translocations by trophectoderm biopsy and comprehensive chromosomal screening. J Assist Reprod Genet. 2018;35:165–9.
Snider AC, Darvin T, Spor L, Akinwole A, Cinnioglu C, Kayali R. Criteria to evaluate patterns of segmental and complete aneuploidies in preimplantation genetic testing for aneuploidy results suggestive of an inherited balanced translocation or inversion. F S Rep. 2021;2:72–9.
Arsham MS, Barch MJ, Lawce HJ. The AGT cytogenetics laboratory manual. John Wiley & Sons; 2017. Available from: https://onlinelibrary.wiley.com/doi/book/10.1002/9781119061199 .
McGowan-Jordan J, Simons A, Schmid M, editors. ISCN 2016: an international system for human cytogenomic nomenclature. S. Karger AG; 2016 [cited 2022 Jun 22]. Available from: https://www.karger.com/Book/Home/271658.
Geiersbach KB, Gardiner AE, Wilson A, Shetty S, Bruyère H, Zabawski J, et al. Subjectivity in chromosome band-level estimation: a multicenter study. Genet Med. 2014;16:170–5.
Yan J, Qin Y, Zhao H, Sun Y, Gong F, Li R, et al. Live birth with or without preimplantation genetic testing for aneuploidy. N Engl J Med. 2021;385:2047–58.
Gardner DK, Lane M, Stevens J, Schlenker T, Schoolcraft WB. Reprint of: Blastocyst score affects implantation and pregnancy outcome: towards a single blastocyst transfer. Fertil Steril. 2019;112:e81–4.
Dong Z, Zhao X, Li Q, Yang Z, Xi Y, Alexeev A, et al. Development of coupling controlled polymerizations by adapter-ligation in mate-pair sequencing for detection of various genomic variants in one single assay. DNA Res. 2019;26:313–25.
Dong Z, Chau MHK, Zhang Y, Yang Z, Shi M, Wah YM, et al. Low-pass genome sequencing-based detection of absence of heterozygosity: validation in clinical cytogenetics. Genet Med. 2021;23:1225–33.
Wang L, Shen J, Cram DS, Ma M, Wang H, Zhang W, et al. Preferential selection and transfer of euploid noncarrier embryos in preimplantation genetic diagnosis cycles for reciprocal translocations. Fertil Steril. 2017;108:620-627.e4.
Reig A, Franasiak J, Scott RT, Seli E. The impact of age beyond ploidy: outcome data from 8175 euploid single embryo transfers. J Assist Reprod Genet. 2020;37:595–602.
Shao L, Shaw CA, Lu X-Y, Sahoo T, Bacino CA, Lalani SR, et al. Identification of chromosome abnormalities in subtelomeric regions by microarray analysis: a study of 5,380 cases. Am J Med Genet A. 2008;146A:2242–51.
Moog U, Arens YHJM, van Lent-Albrechts JCM, Huijts PEA, Smeets EEJ, Schrander-Stumpel CTRM, et al. Subtelomeric chromosome aberrations: still a lot to learn. Clin Genet. 2005;68:397–407.
Menten B, Maas N, Thienpont B, Buysse K, Vandesompele J, Melotte C, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J Med Genet. 2006;43:625–33.
Rivera H, Sitch FL, Crolla JA. Telomeric translocations are uncommon. Genet Couns. 1995;6:343–7.
Shaffer LG, Spikes AS, Macha M, Dunn R. Identification of a subtle chromosomal translocation in a family with recurrent miscarriages and a child with multiple congenital anomalies. A case report. J Reprod Med. 1996;41:367–71.
Wakui K, Tanemura M, Suzumori K, Hidaka E, Ishikawa M, Kubota T, et al. Clinical applications of two-color telomeric fluorescence in situ hybridization for prenatal diagnosis: identification of chromosomal translocation in five families with recurrent miscarriages or a child with multiple congenital anomalies. J Hum Genet. 1999;44:85–90.
Brackley KJ, Kilby MD, Morton J, Whittle MJ, Knight SJ, Flint J. A case of recurrent congenital fetal anomalies associated with a familial subtelomeric translocation. Prenat Diagn. 1999;19:570–4.
Bacino CA, Kashork CD, Davino NA, Shaffer LG. Detection of a cryptic translocation in a family with mental retardation using FISH and telomere region-specific probes. Am J Med Genet. 2000;92:250–5.
Kilby MD, Brackley KJ, Walters JJ, Morton J, Roberts E, Davison EV. First-trimester prenatal diagnosis of a familial subtelomeric translocation. Ultrasound Obstet Gynecol. 2001;17:531–3.
Benzacken B, Carbillon L, Dupont C, Siffroi JP, Monier-Gavelle F, Bucourt M, et al. Lack of submicroscopic rearrangements involving telomeres in reproductive failures. Hum Reprod. 2002;17:1154–7.
Fan Y-S, Zhang Y. Subtelomeric translocations are not a frequent cause of recurrent miscarriages. Am J Med Genet. 2002;109:154.
Yakut S, Berker-Karauzum S, Simsek M, Zorlu G, Trak B, Luleci G. Telomere-specific fluorescence in situ hybridization analysis of couples with five or more recurrent miscarriages. Clin Genet. 2002;61:26–31.
Joyce CA, Dennis NR, Howard F, Davis LM, Thomas NS. An 11p;17p telomeric translocation in two families associated with recurrent miscarriages and Miller-Dieker syndrome. Eur J Hum Genet. 2002;10:707–14.
Cockwell AE, Jacobs PA, Beal SJ, Crolla JA. A study of cryptic terminal chromosome rearrangements in recurrent miscarriage couples detects unsuspected acrocentric pericentromeric abnormalities. Hum Genet. 2003;112:298–302.
Bruyere H, Rajcan-Separovic E, Doyle J, Pantzar T, Langlois S. Familial cryptic translocation (2;17) ascertained through recurrent spontaneous abortions. Am J Med Genet A. 2003;123A:285–9.
Jalal SM, Harwood AR, Sekhon GS, Pham Lorentz C, Ketterling RP, Babovic-Vuksanovic D, et al. Utility of subtelomeric fluorescent DNA probes for detection of chromosome anomalies in 425 patients. Genet Med. 2003;5:28–34.
Alkuraya FS, Martin CL, Kimonis VE. Recurrent miscarriage in a carrier of a balanced cytogenetically undetectable subtelomeric rearrangement: how many are we missing? Prenat Diagn. 2006;26:291–3.
Monfort S, Martínez F, Roselló M, Badia L, Prieto F, Orellana C. A subtelomeric translocation apparently implied in multiple abortions. J Assist Reprod Genet. 2006;23:97–101.
Wise JL, Crout RJ, McNeil DW, Weyant RJ, Marazita ML, Wenger SL. Cryptic subtelomeric rearrangements and X chromosome mosaicism: a study of 565 apparently normal individuals with fluorescent in situ hybridization. PLoS ONE. 2009;4:e5855.
Durmaz B, Karaca E, Durmaz A, Atik T, Akin H, Cogulu O, et al. Subtelomeric rearrangements in patients with idiopathic intellectual disabilitiy/multiple congenital anomalies and recurrent miscarriages: seven years’ experience. Genet Couns. 2013;24:167–77.
Hajlaoui A, Slimani W, Kammoun M, Sallem A, El Amri F, Chaieb A, et al. Subtelomeric rearrangements in patients with recurrent miscarriage. Int J Fertil Steril. 2018;12:218–22.
Zhang S, Pei Z, Lei C, Zhu S, Deng K, Zhou J, et al. Detection of cryptic balanced chromosomal rearrangements using high-resolution optical genome mapping. J Med Genet. 2022;jmedgenet-2022-108553.
Coonen E, Rubio C, Christopikou D, Dimitriadou E, Gontar J, Goossens V, et al. ESHRE PGT Consortium good practice recommendations for the detection of structural and numerical chromosomal aberrations†. Hum Reprod Open. 2020;2020:hoaa017.
Huang C, Jiang W, Zhu Y, Li H, Lu J, Yan J, et al. Pregnancy outcomes of reciprocal translocation carriers with two or more unfavorable pregnancy histories: before and after preimplantation genetic testing. J Assist Reprod Genet. 2019;36:2325–31.
Gardner RM, Sutherland GR, Shaffer LG. Chromosome abnormalities and genetic counseling. Oxford University Press; 2011. Available from: https://academic.oup.com/book/25214 .
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–64.
Gross M, Mkrtchyan H, Glaser M, Fricke HJ, Höffken K, Heller A, et al. Delineation of yet unknown cryptic subtelomere aberrations in 50% of acute myeloid leukemia with normal GTG-banding karyotype. Int J Oncol. 2009;34:417–23.
Jacquemont M-L, Sanlaville D, Redon R, Raoul O, Cormier-Daire V, Lyonnet S, et al. Array-based comparative genomic hybridisation identifies high frequency of cryptic chromosomal rearrangements in patients with syndromic autism spectrum disorders. J Med Genet. 2006;43:843–9.
Faas BHW, Nillesen W, Vermeer S, Weghuis DO, de Leeuw N, Smits APT, et al. Detection of cryptic subtelomeric imbalances in fetuses with ultrasound abnormalities. Eur J Med Genet. 2008;51:511–9.
Scott RT. Introduction: subchromosomal abnormalities in preimplantation embryonic aneuploidy screening. Fertil Steril. 2017;107:4–5.
Xie P, Liu P, Zhang S, Cheng D, Chen D, Tan Y-Q, et al. Segmental aneuploidies with 1 Mb resolution in human preimplantation blastocysts. Genet Med. 2022;S1098-3600(22)00899-1
Chow JFC, Cheng HHY, Lau EYL, Yeung WSB, Ng EHY. Distinguishing between carrier and noncarrier embryos with the use of long-read sequencing in preimplantation genetic testing for reciprocal translocations. Genomics. 2020;112:494–500.
Yanfei Cheng MM, Yu Q, Ma M, Wang H, Tian S, Zhang W, et al. Variant haplophasing by long-read sequencing: a new approach to preimplantation genetic testing workups. Fertil Steril. 2021;116:774–83.
Kurahashi H, Inagaki H, Ohye T, Kogo H, Tsutsumi M, Kato T, et al. The constitutional t(11;22): implications for a novel mechanism responsible for gross chromosomal rearrangements. Clin Genet. 2010;78:299–309.
Johnson SH, Smadbeck JB, Smoley SA, Gaitatzes A, Murphy SJ, Harris FR, et al. SVAtools for junction detection of genome-wide chromosomal rearrangements by mate-pair sequencing (MPseq). Cancer Genet. 2018;221:1–18.
Acknowledgements
We appreciate Miss Zihan Chen for her invaluable help in sample preparation and all participants involved in this study.
Funding
This study was supported by the National Key R&D Program of China (2021YFC2700604), the Collaborative Research Fund (C4062-21GF), and the National Natural Science Foundation of China (82171648 and 32270678).
Author information
Authors and Affiliations
Contributions
S. L., H. L., Z.-J. C., K. W. C., Z. D., and J. Y. designed the whole study. J. Y. recruited patients. S. L. collected samples and variant verification and data curation. H. Li contributed to FISH validation. Z. D. and K. W. C. completed MPseq, data analysis, and variant interpretation. Y. G., Y. Z., and X. Y. collected and analyzed the PGT results. S. L., H. L., K. W. C., Z. D., and J. Y. wrote the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, S., Li, H., Gao, Y. et al. Identification of cryptic balanced translocations in couples with unexplained recurrent pregnancy loss based upon embryonic PGT-A results. J Assist Reprod Genet 41, 171–184 (2024). https://doi.org/10.1007/s10815-023-02999-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-023-02999-2