Abstract
Purpose
Single-nucleotide polymorphisms (SNPs) in the p53 pathways have shown to play a role in endometrial receptivity and implantation in infertile women undergoing in vitro fertilization (IVF). The present study aimed to assess the influence of these gene variants over pregnancy success through a receptivity model in recipients of egg donation treatments, when factors such as age and quality of the oocytes are standardized.
Methods
A nested case–control study was performed on 234 female patients undergoing their first fresh IVF treatment as recipients of donor oocytes. Genotyping of TP53 Arg72Pro (rs1042522), LIF (rs929271), MDM4 (rs1563828), and USP7 (rs1529916) SNPs in the recipients allowed comparison of allele and genotype frequencies and their association with the IVF treatment outcome.
Results
Grouped by genotypes, patients showed differences in IVF outcomes after the embryo transfer. Arg72Pro (rs1042522) gene variant was associated to changes in implantation and clinical pregnancy rates. The polymorphisms USP7 (rs1529916) and MDM4 (rs1563828) were associated to differential ongoing pregnancy rates and variable miscarriage events, respectively.
Conclusions
This study highlights the association between gene polymorphisms related to P53 function and their influence over IVF reproductive outcomes. Arg72Pro variant may influence early events, as lower implantation rates were found in homozygous for Pro72 allele. By contrast, MDM4 (rs1563828) and USP7 (rs1529916) gene variants were associated with the later maintenance of pregnancy.
Similar content being viewed by others
Introduction
Pregnancy establishment requires important cellular and molecular changes through differentiation and proliferation processes [1]. Several biological processes supporting the embryo-maternal dialogue have been identified; however, the role of many interacting molecules and cells in the female reproductive tract during this critical process is still unknown, and it is not accessible for the clinical management in assisted reproduction technology (ART) treatments. Embryo implantation concerns a particularly exceptional mechanism between two cellular entities with different genetic backgrounds and also ontogenetic states, namely the trophoblast of the blastocyst and the epithelial endometrial cells of the uterine cavity, which interact synergistically for a common physiological purpose. The current research on the role of the endometrium in this synergy has focused mainly on its receptive potential. That research has given some insights about transcriptomic and microbiological conditionants in the recipient that could impair the implantation of the embryo [2, 3]. Endometrial receptivity tests are intended to assess solely the endometrial functional state. Additional strategies approaching genetic variability, such as the investigation of mutations, could provide useful data to track relevant conditions limiting the intercommunication between the endometrium and the embryo, and these could be studied using non-invasive methods.
The P53 family comprises a wide number of hyperconnected transcription factors associated to cellular apoptosis, cell cycle arrest, senescence, and DNA repair. The control of these processes by P53 protein products characterizes their function as a tumor suppressor [4,5,6,7]. Due to the maintenance of genome stability by these processes, P53 is known as “the guardian of the genome” [8]. Previous findings have revealed the implication of P53 in embryo implantation mediated by leukemia inhibitory factor (LIF) [9]. It has been suggested that this role must be a more primitive function than the most well known as a tumor suppressor [1, 10]. The role of TP53 has been highlighted in embryo implantation, as the processes involved require intense transformation of cells that are additionally exposed to a very changing environment. It is thus suggested that a protective role of p53 could be relevant for maintaining the cells’ genomic stability at the embryo-maternal interface [11].
The influence of the P53 family in reproduction has been proposed to be dependent on LIF [12], whose critical role during the embryo-maternal dialogue has been demonstrated [13,14,15]. LIF participates in many diverse biological processes in the uterus involving cell proliferation, differentiation, decidualization, immune response, and vasculature remodeling [16]. Current data suggest that fluctuations on LIF levels in the endometrium could impair the establishment and maintaining of the pregnancy and that elevated and maintained LIF levels may improve endometrial receptivity [16, 17].
Additional gene variants related to the P53 pathway have been associated to reproductive success [18,19,20], and previous studies have assessed their influence over outcome of in vitro fertilization (IVF) treatments, however with conflicting results [18,19,20,21,22,23,24]. The available studies have mainly included patients that used homologous oocytes in IVF treatments; thus, important confounders such as infertility factors related to oocyte biology and female age have made it difficult to disentangle the influence of the gene variants investigated on the treatment outcomes [10].
For this study, we used the model of pregnancy establishment in women undergoing treatments using donor oocytes, to achieve a receptivity model for the investigation of biological features regarding embryo implantation when the recipient does not share genetics with the embryo to be implanted and to minimize the effect of factors intrinsically attained to oocyte competence, such as age.
For this genetic association study, we settled a list of candidate single-nucleotide polymorphisms (SNPs) related to P53 pathway, and these were genotyped to explore the reproductive outcome of IVF treatments in women recipient of donor oocytes. The selected variants consisted of the most known functional variant rs1042522 (Arg72Pro) in TP53; the rs929271 of LIF gene, whose function is essential during embryo implantation and which also seems to regulate P53 [13, 25]; the variant rs1563828 on Murine double minute 4 (MDM4) whose protein product inhibits P53 activity [26]; and the SNP rs1529916 located in the ubiquitin-specific-processing protease 7 (USP7) gene that shows a splicing variability according to genotype [26], as its protein product has also shown to modify P53 levels [27]. All the genes containing such selected variants were also selected as they are expressed in the endometrium robustly and consistently [28].
Material and methods
Study design and patient inclusion
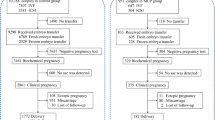
A genetic association study with a nested case–control design was performed to assess the effect of selected candidate gene variants related to P53 pathway on the outcome of oocyte donation treatments and pregnancy progress. Patients undergoing IVF with donated oocytes were recruited for the present study at a private IVF unit. Included women were European Caucasian aged between 18 and 45 years undergoing the first oocyte donation treatment as recipients and receiving a fresh embryo transfer (ET) of at least two good-quality cleavage-stage embryos on day 2 or day 3. Women were excluded if they presented with uterine factor infertility, abnormal endometrial thickness, suspected hydrosalpinx, body mass index (BMI) > 30 kg/m2, or if there was severe male factor infertility in the partner. The patients provided a signed consent form for the IVF treatment and also for the genetic study and provided a DNA sample obtained from buccal swab.
Controlled ovarian hyperstimulation (COH) in oocyte donors, oocyte retrieval, insemination, and embryo culture
Oocyte donors were European Caucasian healthy women with good physical and mental health aged 18–35 years, having regular menstrual cycles and a body mass index between 18 and 29 kg/m2. Donors were screened for known hereditary disorders, chromosome abnormalities, and infectious diseases. Donors were not accepted if they previously presented an abnormal response to gonadotropin, polycystic ovary syndrome (PCOS), endometriosis, more than one previous miscarriage, and any other medical condition that could compromise the success of the treatment.
Human recombinant follicle-stimulating hormone (hrFSH) and gonadotropin-releasing hormone (GnRH) antagonists were used for COH in donors, and hrFSH units were adjusted between 150 and 300 IU depending on age, BMI, antral follicle count (AFC), and ovary response. Oocyte maturation was triggered by administering GnRH analogue after having reached at least three dominant follicles ≥ 17 mm. Oocyte retrieval was done 35 h after the GnRH analogue administration. Denudation of cumulus oocytes complexes was performed by mechanical disaggregation after a brief exposure of hyaluronidase (40 IU/mL) in HEPES. Mature metaphase II oocytes obtained were inseminated using sperm from the male partner through intracytoplasmic sperm injection (ICSI) 2 h after denudation. Embryo culture was performed in low oxygen atmosphere 5% (O2) and 6% (CO2) using sequential commercial culture medium cleavage and blastocyst (SAGE).
Embryo transfer
Embryo transfer (ET) to the recipients was performed on day + 2 or day + 3. Recipients underwent endometrial preparation based on suppression of ovarian function and increasing dosage of estradiol valerate tablets. From the day of donor’s oocyte retrieval, the recipients received micronized progesterone (P) vaginal pessaries for luteal phase support. The ET was performed only after confirmation of an endometrium with typical trilaminar pattern with a thickness of at least 6 mm. Embryos were selected for ET when at least two good-quality embryos reached grade A or B following Spanish Society for Studies on Reproductive Biology (ASEBIR) [29].
Clinical outcomes
Pregnancy was determined by serum or urine tests detecting positive levels of human choriogonadotropin hormone β-hCG in oocyte recipients after two consecutive measurements in day + 11 or + 12 after ET, depending on cleavage stage of transferred embryos day + 3 or + 2, respectively (β-hCG ( +)). Clinical pregnancy (CP) was determined by ultrasonographical evidence of a gestational sac 5–6 weeks after ET. The absence of an ultrasound-identifiable pregnancy and a decrease in the levels of β-hCG represented a biochemical pregnancy loss (BPL). An ongoing pregnancy (OP) was defined as the presence of at least one viable fetus after week 12 of gestation. Miscarriages (M) were considered to be those gestational losses that occurred between weeks 4 and 20, having previously identified a gestational sac [30]. The implantation rate (IR) was calculated as the ratio between the number of gestational sacs found by ultrasound scans and the number of embryos transferred. Ectopic pregnancy was defined as those ultrasound observations of a sac outside the uterus [31]. Those recipients with no evidence of pregnancy after the treatment were defined as the nonpregnant group.
DNA preparation and genotyping
Genomic DNA was isolated from buccal swab collected from the recipients and using the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA). Genotyping process was outsourced to the genetic and proteomic services of Science and Technology School in the University of Pais Vasco (Bizkaia). TaqMan® Open Array Genotyping System (Applied Biosystems, UK) was used to identify TP53 (rs1042522), LIF (rs929271), MDM4 (rs1563828), and USP7 (rs1529916). Allelic assignment was done using TaqMan Genotyper® (Applied Biosystems, UK). Positive and negative controls were included in each reaction assay to ensure the quality and concordance of results to validate them. A minimal calling rate of 80% was used to assign genotype in the SNP. The list of gene variants included in the panel is summarized in Table 1.
Statistical analysis
The study was designed as a nested case–control in which cohort groups are established according to the results obtained after the IVF treatment. This enables the possibility of analyzing the incidence of the genotypes according to the population stratified by treatment results and by pregnancy progress. Positive or favorable outcome was settled as control and negative outcome as a case.
To assess any possible population bias compared to the general population, allele and genotype frequencies published for the studied SNPs in 1000 Genomes (Phase 3) were used as reference [32]. Differences between allele and genotype frequencies of 1000 Genomes population and the recipients, object of the study, were calculated by using chi-squared test that was also used to assess compliance of Hardy–Weinberg equilibrium between genotype frequencies of every SNP on each group. Pooled by genotype, differences between age and number of embryos transferred were calculated by analysis of the variance (ANOVA). No multiple test adjustment was carried out as the selection of SNPs was based on previous association studies.
The clinical treatment outcomes IR, CP, BPL, M, and OP were used to assess the treatment progression and also their variation between recipients grouped by genotypes through Fisher’s exact and chi-squared tests. When a variation in the number of recipients was present on any group representing a clinical significance, a logistic regression model was used to compute the odds ratio with a 95% confidence interval, and the age of the recipient was used as covariate and treated as a potential confounder for adjustment. Groups of genotypes for comparison were established by considering the possible recessive, dominant, codominant, and over-dominant models of inheritance. To provide the fittest model of interaction between genotypes, and assuming a total absence of a known direct biological influence, Akaike’s (AIC) and Bayesian’s (BIC) information criteria were used to select the degree of association for every genotype with treatment outcomes [33]. Statistical significance was considered when p < 0.05 2-sided for all the tests.
Results
A total of 234 Caucasian European recipients undergoing an IVF treatment with donated oocytes aged 40.48 (± 4.37) were included in the study (Table 2). Most women attempted oocyte donation treatment due to advanced reproductive age (57.0%), ovarian failure (23.3%), previous poor response to gonadotropins in IVF attempts (12.5%), endometriosis affecting the ovary (4.0%), and unspecified reasons (3.2%). Assessment of β-hCG in blood or urine test performed 14 days after oocyte insemination allowed dividing the cohort into two groups: a total of 150 recipient had a positive β-hCG ( +), and the remaining 84 had a negative result β-hCG ( −) (Table 2).
Allelic and genotype frequency distribution for the studied gene variants did not vary between the group of patients and the reference population (Caucasian European 1000 Genomes, Phase 3 GRCh37, public data), and the only differences were observed for the MDM4 gene variant (rs1563828) (Table 3). The two groups β-hCG ( +) and β-hCG ( −) were compared for every SNP to assess deviation of Hardy–Weinberg (HW) equilibrium, and a significant variation between genotype frequencies was observed for the MDM4 variant in the non-pregnant recipients (p = 0.006) (Table 3).
The distribution of allele variants for the allele Pro72 (rs1042522) (C) showed a significant 12% increase in recipients tested β-hCG ( −) (p = 0.016) compared to β-hCG ( +), which resulted in an increase of homozygous Pro72 (C/C) recipients in the non-pregnant recipients, compared with the β-hCG ( +) group (Table 3). A significant increase of the A/A homozygous genotype of the USP7 (rs1529916) gene variant was observed in β-hCG ( +) patients (Table 3). All the remaining SNPs showed no variation in allele and genotype frequency distribution between these two groups of patients (Table 3).
IVF treatment outcomes are presented individually per genotype in (Table 4). A negative trend for homozygous Pro72 recipients was observed by decreased implantation and clinical pregnancy rates (Table 4). Homozygous Pro72 recipients presented almost halved IR (21.95%) compared with heterozygous Arg72/Pro72 (42.34%) and homozygous Arg72 (39.31%) (p = 0.047); in the same line, these recipients also presented significantly reduced CP (38.89%) compared with the Arg72/Pro72 heterozygous (58.67%) and the Arg72 homozygous (68.42%). Recipients grouped by genotypes of the MDM4 (rs1563828) and USP7 (rs1529916) presented differences in the miscarriage and also in OP rates, respectively (Table 4).
Logistic regression analysis showed that recipients that were homozygous for Pro72 had a higher probability of not achieving pregnancy, and according to AIC and BIC parameters, the recessive model for the Pro72 allele fitted with better score between the other evaluated models (Table 5). The A allele of USP7 (rs1529916) was associated to increased OP under a dominant model (Table 5). The over-dominant model fitted better for MDM4 (rs1563828), and heterozygous A/G women presented with fewer miscarriages (Table 5).
Discussion
The present study used a model based on donated oocytes to investigate the association between p53-related gene variants and the success of IVF treatments in women recipients of donor eggs. Our results showed that TP53 Arg72Pro (rs1042522) was associated to higher likelihood of achieving successful embryo implantation and pregnancy establishment. Our study suggests that certain clinical outcomes could be associated to USP7 (rs1529916) and MDM4 (rs1563828) gene polymorphisms, which might affect the following events for maintenance of an ongoing pregnancy.
The group of recipients that did not achieve pregnancy showed a significant increase in the frequency of the allele Pro72 (Table 3), and IR were also decreased by half in recipients homozygous for this allele, which was associated with significant lower CP rates compared to recipients carrying at least one Arg72 allele (Table 4). USP7 gene variant (rs1529916) was associated with later pregnancy events, and recipients carrying at least one A allele had higher OP rates than homozygous G/G. However, it is very interesting that the homozygous genotype A/A, which represents the lowest frequency not only among the studied population but also in the wide European people (1000 Genomes) [32], significantly increased in the group of recipient β-hCG ( +) (Table 3) and also presented with a trend to higher IR compared to the genotypes A/G and G/G, although this comparison did not reach statistical significance (Table 4). These data suggest that the USP7 variant might also be associated with earlier events during pregnancy establishment and not only with events in the later stages. A lower risk for miscarriage was observed for heterozygous of the MDM4 gene variant. These results reinforce the idea about the potential utility of p53-related gene variants as predictive markers in IVF and more specifically as an indicator of receptivity potential.
In this study, we did not screen for abnormal chromosomal errors in the embryos, which is a limitation of the study; however, to minimize this potential impact, our study was designed using the model of egg donor treatment, where donors are young and healthy women. Our results are in agreement with previous genetic association studies reporting a negative impact of Pro72 homozygous genotype on IVF treatments outcomes [18,19,20, 22, 24]. It is striking that IR varied between patients grouped by genotypes following a very similar trend in our study, in comparison to what has been observed in these previous reports. Variation of IR has been almost halved not only in women homozygous for Pro72 allele that were young (< 35 years) [19], but also in slightly older patients (37.9 ± 3.3 years) that were recruited due to previous history of recurrent implantation failure [20].
The available literature on possible associations between p53 gene variants and IVF outcome have so far had little or null impact in the practice of reproductive medicine. A main reason may be dependent on the previous inconsistent results reported [21] or that some studies have found an association between the Pro72 allele and a better reproductive outcome of IVF treatments [23]. A possible explanation is that the effect of the Arg72Pro variant seems to be moderate, and it may have gone unnoticed in both these previous studies due to recruitment of patients with various causes of infertility and a wide age range, both factors that could have both impacted oocyte competence. The recruitment of heterogeneous populations could additionally have masked the effect of these gene variants in the previous studies [10].
Our study aimed at investigating the impact of these gene variants of p53 on IVF outcomes, however using a specific model with good expected oocyte competence from donors and reduced dependency of female age factors, where the model of implantation and pregnancy establishment would be less impacted by these bias. Studies on IVF treatments in oocyte recipients have been previously conducted, but little information has been provided regarding the inclusion criteria of recipients and the selection criteria and age limit of the oocyte donors [19]. The recipients of our study were aged 40.48 (± 4.37) years, and the IVF outcomes varied according to genotypes similarly to the previous studies using homologous oocytes [18,19,20]; in addition, donors were aged within 18–35 years, and the influence of oocyte competence as a possible bias is expected to be decreased; hence, we could hypothesize that Arg72Pro variant may be associated with endometrial receptivity independently of the oocyte competence.
The genes MDM4 and USP7 are highly expressed in the endometrium, and they are critical regulators affecting P53 activity [28, 34, 35]. Although the functional impact of both selected intronic SNPs is unclear, they were also associated to infertile population in previous reports [19]. The data on association between (rs1563828) MDM4 variant and lack of pregnancies has to be interpreted with caution, as it could be due to induced bias, as it did not accomplish HW equilibrium in the recipients that did not achieve pregnancy.
Biological processes by which p53 could mediate embryo implantation remain unknown. LIF has been postulated as a mediator of the p53 pathway, and it has been shown to regulate embryo implantation in mammals [36,37,38]; however, its role in pregnancy maintenance is not clear [14]. A functional implication of Arg72Pro has been widely studied, and previous experiments showed that the apoptotic activity associated to p53 is less efficient when the allele Pro72 is present [39]. A limitation of this apoptotic function could be a potential path by which Pro72 allele could reduce the pregnancy success as this role is needed for dialogue between the implanting blastocyst and receptive endometrium [40, 41]. Quantification of LIF expression indicates that it is two times higher in Arg72 homozygous carriers compared to that in Pro72 homozygous, which could impact differently success of pregnancy establishment [9]. Our results fitted with these previous observations, and the recessive model of the allele Pro72 is supported by these biological data. The higher IR in recipients carrying Arg72, of about twice that of recipients homozygous for Pro72, supports also the data on LIF functional expression that has been estimated for these genotypes [19]. Our results thus support the hypothesis that homozygous Pro72 recipients could present diminished LIF levels, and this could impair cellular and molecular dialogue during early embryo implantation, however without clear influence on the later progress of the pregnancy. By contrast, USP7 (rs1529916) and MDM4 (rs1563828) gene variants may have an impact in later stages. At present, the functional implications of these intronic changes are unknown, but it could be hypothesized that perhaps they could be linked to any other functional variant or at least mediate, under a genomic regulatory mechanism that potentially modify p53 activity and/or levels, in a manner that may impair the pregnancy maintenance.
Our data add to the current literature and our observations merit further investigation of the influence of p53 on embryo implantation and evaluate its predictive value for the clinical management of patients undergoing IVF treatments. As proposed by previous authors, controlled therapies based on exogenous administration of LIF or modification of p53 levels and/or activity could be a potential alternative to explore new research or future clinical improvements [10].
The present study provides support of a clinical association between polymorphisms located at genes involved in the P53 pathway and the success of IVF in recipients of donor oocytes, which was used as a model of pregnancy establishment independent of the oocyte genetic origin and reducing the bias of poor oocyte competence. Further studies are needed to clarify the biological context of our findings and to extend the knowledge of polymorphism of genes of the P53 pathway in different populations and ethnicities.
References
Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18(12):1754–67.
Moreno I, Codoñer FM, Vilella F, Valbuena D, Martinez-Blanch JF, Jimenez-Almazán J, et al. Evidence that the endometrial microbiota has an effect on implantation success or failure. American Journal of Obstetrics & Gynecology. Elsevier; 2016;215:684–703.
Ruiz-Alonso M, Blesa D, Díaz-Gimeno P, Omez EG, Fern Andez-S Anchez M, Carranza F, et al. The endometrial receptivity array for diagnosis and personalized embryo transfer as a treatment for patients with repeated implantation failure. Fertility and Sterility. 2013;100:818–24.
Hu W, Feng Z. The role of p53 in reproduction, an unexpected function for a tumor suppressor. Journal of Molecular Cell Biology. Oxford University Press; 2019. p. 624–7.
Kung CP, Murphy ME. The role of the p53 tumor suppressor in metabolism and diabetes. Journal of Endocrinology. BioScientifica Ltd.; 2016. p. R61–75.
Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. Cell; 2009. p. 413–31.
Feng Z, Levine AJ. The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends in Cell Biology. Trends Cell Biol; 2010. p. 427–34.
Lane DP. p53, guardian of the genome. Nature. Nature; 1992. p. 15–6.
Hu W, Feng Z, Teresky AK, Levine AJ. p53 regulates maternal reproduction through LIF. Nature. 2007;450:721–4.
Kang HJ, Rosenwaks Z. p53 and reproduction. Fertility and sterility. Elsevier Inc.; 2018. p. 39–43.
Hu W. The role of p53 gene family in reproduction. Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor Laboratory Press; 2009;1.
Feng Z, Zhang C, Kang H-J, Sun Y, Wang H, Naqvi A, et al. Regulation of female reproduction by p53 and its family members. FASEB J. 2011;25:2245–55.
Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Köntgen F, et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359:76–9.
Chen JR, Cheng J-G, Shatzer T, Sewell L, Hernandez L, Stewart CL. Leukemia inhibitory factor can substitute for nidatory estrogen and is essential to inducing a receptive uterus for implantation but is not essential for subsequent embryogenesis 1. Endocrinology. 2000;141:4365–72.
Ni H, Ding N-Z, Harper MJK, Yang Z-M. Expression of leukemia inhibitory factor receptor and gp130 in mouse uterus during early pregnancy. Mol Reprod Dev. 2002;63:143–50.
Mariee N, Li TC, Laird SM. Expression of leukaemia inhibitory factor and interleukin 15 in endometrium of women with recurrent implantation failure after IVF correlation with the number of endometrial natural killer cells Human reproduction (Oxford, England). Oxford University Press. 2012;27:1946–54.
Rosario GX, Stewart CL. The multifaceted actions of leukaemia inhibitory factor in mediating uterine receptivity and embryo implantation. Am J Reprod Immunol. 2016;75:246–55.
Kay C, Jeyendran RS, Coulam CB. p53 tumour suppressor gene polymorphism is associated with recurrent implantation failure. Reprod Biomed Online. 2006;13:492–6.
Kang HJ, Feng Z, Sun Y, Atwal G, Murphy ME, Rebbeck TR. Single nucleotide polymorphisms in the p53 pathway regulate fertility in humans. Proceedings of the National Academy of Sciences of the United States of America. National Academy of Sciences. 2009;106:9761–6.
Lledo B, Turienzo A, Ortiz JA, Morales R, Ten J, Llácer J, et al. Negative effect of P72 polymorphism on p53 gene in IVF outcome in patients with repeated implantation failure and pregnancy loss. J Assist Reprod Genet. 2014;31:169–72.
Patounakis G, Treff N, Tao X, Lonczak A, Scott RT, Frattarelli JL. The p53 codon 72 single nucleotide polymorphism lacks a significant effect on implantation rate in fresh in vitro fertilization cycles: an analysis of 1,056 patients. Fertil Steril. 2009;92:1290–6.
Paskulin DD, Cunha-Filho JSL, Souza CAB, Bortolini MC, Hainaut P, Ashton-Prolla P. TP53 PIN3 and PEX4 polymorphisms and infertility associated with endometriosis or with post-in vitro fertilization implantation failure. Cell Death and Disease. 2012;
Chan Y, Zhu B, Jiang H, Zhang J, Luo Y, Tang W. Influence of TP53 codon 72 polymorphism alone or in combination with HDM2 SNP309 on human infertility and IVF outcome. PloS one Public Library of Science. 2016;11:e0167147.
Mohammadzadeh M, Ghorbian S, Nouri M. Evaluation of clinical utility of P53 gene variations in repeated implantation failure. Molecular Biology Reports. Springer, Netherlands. 2019;46:2885–91.
Yu H, Yue X, Zhao Y, Li X, Wu L, Zhang C, et al. LIF negatively regulates tumour-suppressor p53 through Stat3/ID1/MDM2 in colorectal cancers. Nature Communications Nature Publishing Group. 2014;5:5218.
Ohtsubo C, Shiokawa D, Kodama M, Gaiddon C, Nakagama H, Jochemsen AG, et al. Cytoplasmic tethering is involved in synergistic inhibition of p53 by Mdmx and Mdm2. Cancer Science. 2009;
Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004.
Thul P, Åkesson L, Wiking M, Mahdessian D, Geladaki A, Ait Blal H, et al. The human protein Atlas. 2020.
Cortés JL, Ligero G, Sánchez L, Nieto Ruiz de Zárate AI, Bueno C, Montes Ramírez MIR, et al. Criterios de valoración morfológicos de oocitos, preembriones tempranos y blastocistos humanos propuestos por ASEBIR. ASEBIR, ISSN-e 1136–4424, Vol 13, No 1, 2008. Asociación para el Estudio de la Biología de la Reproducción. 2008;13:2.
Farquharson RG, Jauniaux E, Exalto N. Updated and revised nomenclature for description of early pregnancy events. Human reproduction (Oxford, England). 2005;20:3008–11.
Kolte AM, on behalf of the ESHRE Special Interest Group EP, Bernardi LA, on behalf of the ESHRE Special Interest Group EP, OB Christiansen on behalf of the ESHRE Special Interest Group EP, et al. Terminology for pregnancy loss prior to viability a consensus statement from the ESHRE early pregnancy special interest group. Human Reproduction. Oxford Academic. 2015;30:495–8.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature Nature Publishing Group, a division of Macmillan Publishers Limited. All Rights Reserved. 2012;491:56–65.
Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics (Oxford, England). 2006;22:1928–9.
Stad R, Little NA, Xirodimas DP, Frenk R, van der Eb AJ, Lane DP, et al. Mdmx stabilizes p53 and Mdm2 via two distinct mechanisms. EMBO reports. John Wiley & Sons, Ltd. 2001;2:1029–34.
Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 2002 416:6881. Nature Publishing Group. 2002;416:648–53.
Cambra JM, Jauregi-Miguel A, Alvarez-Rodriguez M, Parrilla I, Gil MA, Martinez EA, et al. Allogeneic embryos disregulate leukemia inhibitory factor LIF and its receptor in the porcine endometrium during implantation. Frontiers in Veterinary Science Frontiers. 2020;0:993.
Sengupta J, Lalitkumar PGL, Najwa AR, Ghosh D. Monoclonal anti-leukemia inhibitory factor antibody inhibits blastocyst implantation in the rhesus monkey. Contraception Elsevier. 2006;74:419–25.
Winship A, Correia J, Zhang J-G, Nicola NA, Dimitriadis E. Leukemia inhibitory factor (LIF) inhibition during mid-gestation impairs trophoblast invasion and spiral artery remodelling during pregnancy in mice. PLOS ONE Public Library of Science. 2015;10:e0129110.
Dumont P, Leu JI-J, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nature Genetics. 2003;33:357–65.
Welsh AO. Uterine cell death during implantation and early placentation. Microscopy Research and Technique. 1993;
Galán A, O’Connor JE, Valbuena D, Herrer R, Remohí J, Pampfer S, et al. The human blastocyst regulates endometrial epithelial apoptosis in embryonic adhesion1. Biology of Reproduction. 2000;63:430–9.
The Genotype-Tissue Expression (GTEx).
Funding
Open access funding provided by Karolinska Institute. This work was supported by grant PTQ 09–01-00496, Ministerio de Economía y Competitividad (MINECO), Government of Spain, Instituto de Fertilidad Clínica Rincón y Fundación Rincón. Dr AR Palomares and Dr KA Rodriguez-Wallberg are supported by grants from the Swedish Research Council (Dnr 2020–02230), the Swedish Cancer Society (CAN 2017/704, 20 0170 F), and the Karolinska Institutet Research grants.
Author information
Authors and Affiliations
Contributions
Conceptualization, ARP and ARE; methodology, ARP, AACD, and MRG; formal analysis and investigation, ARP, KRW, and ARE; writing (original draft preparation), ARP; writing (review and editing), ARE and KRW; funding acquisition, ARP, ARE, and KRW.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The patients provided a signed consent form for the IVF treatment and also for the genetic study. The study was approved by the ethics committee of the Hospital Regional Universitario Virgen de la Victoria, and the study was carried out in collaboration between the University of Malaga and Instituto de Fertilidad Clínica Rincón.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Palomares, A.R., Castillo-Domínguez, A.A., Ruiz-Galdón, M. et al. Genetic variants in the p53 pathway influence implantation and pregnancy maintenance in IVF treatments using donor oocytes. J Assist Reprod Genet 38, 3267–3275 (2021). https://doi.org/10.1007/s10815-021-02324-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10815-021-02324-9