Abstract
The green alga Ulva ohnoi M. Hiraoka & S. Shimada is an effective biofiltration agent of fish effluents in Integrated Multi-Trophic Aquaculture Recirculation Systems (IMTA-RAS), due to its high growth rate and high nutrient (N and P) removal in temperate conditions. Moreover, Ulva species provide an important niche for biofilm-forming bacteria, including strains of Phaeobacter that possess antagonistic activity against pathogenic Vibrio species, and have been shown to reduce the mortality of fish in aquaculture settings. This research aimed to examine the potential of using U. ohnoi colonised with Phaeobacter sp. 4UAC3, previously isolated from natural populations of Ulva australis, as a strategy to prevent vibriosis in IMTA-RAS. The results showed that Phaeobacter sp. 4UAC3 was able to colonise and be maintained on U. ohnoi during a re-inoculation scale-up process from multi-well plates (10 mL) to flasks (4 L) and finally to tanks (40 L). Phaeobacter sp. 4UAC3 rapidly colonised the surface of the Ulva but it did not significantly modify the rest of the bacterial communities present in U. ohnoi surface in terms of diversity and composition. Infection assays using fish larvae (Scophthalmus maximus) with V. anguillarum in a model IMTA-RAS showed that Phaeobacter–colonized U. ohnoi promoted the reduction of mortality in the infected larvae, however this trend was not statistically supported. These results suggested that U. ohnoi can be experimentally colonised with Phaeobacter sp. 4UAC3 and has potential to be used in IMTA-RAS as an alternative to traditional disease control methods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The use of Ulva (Ulvophyceae, Chlorophyta) in the biofiltration of fish effluents in Integrated Multi-Trophic Aquaculture (IMTA) systems is of growing interest, primarily due to their high surface/volume ratio that allows them to have high nutrient uptake rates, especially in environments rich in N (mainly NO3− and NH4+) and P (PO43−). Other characteristics that make Ulva a good candidate for use in IMTA systems include their high growth rate, their capacity of vegetative reproduction and to be cultured unattached, as well as their ecophysiological plasticity, being well adapted to a wide range of environmental conditions (Lawton et al. 2013; Bolton et al. 2016; Cremades et al. 2017). In addition, these algae are gaining ground as raw materials in food and feed, for the production of bioactive compounds in functional foods or the production of biofuels (Mantri et al. 2020). Among several species tested in previous studies, Ulva ohnoi M. Hiraoka & S. Shimada was selected as an ideal candidate for the filtration of Senegalese sole (Solea senegalensis) culture effluents in an IMTA Recirculation Aquaculture System (IMTA-RAS), due to be adapted to a growth temperature which fits well Senegalese sole production (Oca et al. 2019).
Bacterial communities associated to Ulva spp. are different from those of the surrounding water both in terms of biodiversity and function, and play an essential role for the growth and development of the algae, shaping key phenotypes (e.g., morphology, (Wichard et al. 2015)) or having a role in algae nutrition and defence (Egan et al. 2013; Hollants et al. 2013). These facts give strong support to the notion that macroalgae and all their associated microbiota form a singular functional entity or holobiont (Egan et al. 2013). Bacterial communities of Ulva species from different geographic locations were found to be too variable to describe a community core based on their taxonomic composition, however specific functional genes were enriched across Ulva samples, suggesting the existence of a stable core set of functions provided by bacteria with distinct phylogenetic origin (Roth-Schulze et al. 2018).
Moreover, Ulva laminar thallus provides an important niche for biofilm-forming bacteria, including strains of Phaeobacter that reduce growth and kill fish pathogens, such as Vibrio (D’Alvise et al. 2013) and Tenacibaculum (Tesdorpf et al. 2022) species, possibly due to the production of antibiotic tropodithietic acid (TDA, (Brinkhoff et al. 2004)). It has been demonstrated that Phaeobacter bacteria are effective as probiotic in aquaculture by reducing the mortality of vibrio-challenged fish larvae, administrated bioensulated in the live prey (Planas et al. 2006) or immobilized in biofilters (Prol-García and Pintado 2013). Therefore, using Ulva as a substrate for probiotic bacteria (such as Phaeobacter) could be a good sustainable microbial control strategy in IMTA-RAS. This approach would be in line with ecological theories that promote the resilience of aquaculture systems rather than eradicating pathogens (de Schryver and Vadstein 2014; Egan and Gardiner 2016).
This research aims to examine the potential of using U. ohnoi colonised with Phaeobacter sp. 4UAC3, a bacterium previously isolated from natural populations of Ulva australis Areschoug in Galicia (NW Spain, unpublished), as a strategy to prevent vibriosis in fish cultured in IMTA-RAS. The first objective was to determine the feasibility of co-cultivating U. ohnoi with Phaeobacter. Specifically, we studied the colonization and maintenance of Phaeobacter on U. ohnoi during the scaling-up process of the algae culture (10 mL plate—4 L flask—40 L tank). In addition, the effect of the presence of Phaeobacter on the growth of Ulva and epiphytic microbial communities was analysed. A second objective was to test the probiotic effect of U. ohnoi colonised by Phaeobacter. For this purpose, an infection assay was performed with fish larvae (Scophthalmus maximus) infected with Vibrio anguillarum in a miniaturised IMTA-RAS.
Materials and methods
Two sets of experiments were performed: 1) Scale-up of the co-culture of Ulva and Phaeobacter, where colonization and maintenance of Phaeobacter sp. in Ulva ohnoi and water was studied. In this experiment Ulva growth rate was assessed. 2) In-vivo antagonism assay with fish larvae, where the probiotic effect of U. ohnoi colonized by Phaeobacter was studied. In this experiment, mortality of larvae and permanence of Phaeobacter in Ulva and water were assessed.
Experimental designs
Scale-up of the co-culture of Ulva ohnoi and Phaeobacter sp.
For the scale-up study of Ulva culture with Phaeobacter, two conditions were tested: Ulva-Control (UC), without the addition of Phaeobacter, and Ulva-Phaeobacter (UP). Briefly, 120 algal discs of 2 cm of diameter were grown in multi-well plates. Half of the plates were inoculated with Phaeobacter at a concentration of 107 CFU mL−1 (hereon in referred to as UP), the second half were left uninoculated and used as controls (hereon in referred to as UC). Plates were maintained for 7 days in an incubator (New Brunswick Scientific Excella E24r Temperature-Controlled Benchtop Shaker) with temperature regulation at 18 ºC, orbital shaking at 80 rpm and Daylight-type LED Panel Surface SAMSUNG 44W, 6000 K and 3960 Lm (115 μmol photons m−2 s−1) and 12L:12D photoperiod. During this period, algae disc samples were taken in duplicates at time 0, 2 and 7 (days) for algal growth, microbiological and water physicochemical analyses. Plates were numbered and samples were drawn randomly, using the random.org server to generate random numbers (https://www.random.org).
On day 7, the discs from the plates were transferred to four 5 L balloon flasks (17 discs per flask) with 4 L of culture medium and Phaeobacter was re-inoculated in the UP condition at a concentration of 107 CFU mL−1. The flasks were maintained with aeration, in an incubator cabinet with temperature regulation at 19 °C and daylight-type LED tube 6000 K lighting (170 µmol photons m−2 s−1) with 16L:08D photoperiod for 7 days. At day 14, two thalli were taken from each condition for algal growth, microbiological and physicochemical analyses.
On day 14, the entire algal biomass (~ 15–20 g wet weight) from each flask was transferred to four opened tanks with 40 L of culture medium. Phaeobacter was re-inoculated in the UP thanks at a concentration of 107 CFU mL−1. The tanks were kept with aeration, in a thermoregulated room at 20 °C with daylight-type LED panel 6000 K (275 µmol photons m−2 s−1) with 12L:12O photoperiod for 21 days more. At day 21 and 28, Ulva biomass was collected, spun, weighted and restocked in the tanks with fresh culture medium. At day 28, Ulva biomass was restocked at 120 g per tank to maintain stocking density at similar levels (around 3 kg m−3).
Samples corresponding to days 7, 14, 21, 28 and 35 of Ulva surface microbiota DNA were chosen to sequence the 16S rRNA gene, in order to identify the composition of the microbial communities on the surface of the algae.
In-vivo antagonism assay with fish larvae
To test the probiotic capacity of U. ohnoi colonised with Phaeobacter sp., an infection challenge was performed with turbot (Scophthalmus maximus) larvae based on the procedure described in Planas et al. (2005) and adapted to a miniaturised IMTA-RAS with Ulva control (UC) and with Ulva colonised with Phaeobacter sp (UP) together with a control without Ulva (C). In all three situations, larvae were infected with Vibrio anguillarum through live feed (rotifers) and compared to the same conditions without infection. Therefore, a total of six conditions were studied: Control (C), Ulva-Control (UC), Ulva-Phaeobacter (UP), Control-Vibrio (CV), Ulva-Control-Vibrio (UCV) and Ulva-Phaeobacter-Vibrio (UPV).
The miniaturised IMTA-RAS (Fig. 1) consisted of a 2.8-L tank (Standard M3 Tank, Aquaneering Inc, USA) for larvae (Larvae Tank); connected in a recirculation system to a 3-L tank for algae (Algae Tank), making a total volume of 5.8 L. The overflow from the larvae tank flowed into the algae tank, both separated by a 500 μm mesh. The algae water was pumped back into the larvae tank at a rate of 10 mL min−1. A screen was placed between the two tanks to independently establish the light intensity and photoperiod conditions suitable for the culture of fish larvae and algae (described below). The set-up consisted of 12 systems in parallel, two for each condition studied.
Schematic diagram of the miniaturised IMTA-RAS system. The small-scale system simulating an IMTA-RAS consisted of two connected tanks, one with algae (Ulva ohnoi) and the other with turbot larvae (Scophthalmus maximus). Each tank had an aeration system, and the filtered seawater was recirculated between them by a peristaltic pump. A 500 µm mesh separated the two tanks to prevent the larvae from being transferred to the algae tank. The algae tank was illuminated with a daylight LED tube
The experiment lasted a total of 7 days. From day 0, when 3-days old larvae started to be fed with rotifers, a daily larval mortality count was started. On day 1, samples of water and algal discs were taken for microbiological analysis in duplicates. Further microbiological analyses were done at 6 days for water and 7 days for algae.
Ulva ohnoi culture
Ulva ohnoi were obtained from a single sporophyte culture maintained in Centro Interdisciplinar de Química y Biología (CICA, Universidade da Coruña, Galicia, Spain). The clone (sporophyte) was originally collected at the Parque Natural de las Marismas del Odiel, Huelva, Spain (latitude, 37º14′ N; longitude, 6º59′ W, October 17, 2015) and genetically identified by DNA extraction and PCR amplification of the chloroplast rbcl gene following the protocol described in Hayden et al. (2003) with the primers used by Manhart (1994). It was maintained in “tumble culture” with aeration from the bottom of the tanks, with a weekly change of culture medium and the density adjusted to 3 kg m−3.
In order to improve colonisation of Phaeobacter inoculum by reducing the number of bacteria on the surface of the algae, the Ulva blades were immersed in sterile seawater containing chlorine (1 mL L−1), and then rinsed 3 times with sterile natural seawater (SSW, autoclaved at 121 °C 20 min). The algae blades were punched under sterile conditions to obtain 2 cm diameter discs (3.14 cm2 surface), preferentially selecting non-marginal areas. The resulting discs were washed three times with SSW and distributed in 6-well plates (Corning. diameter 34.8 mm, volume 16.8 mL, area 9.5 cm2), one for each well, filled with 10 mL of Ulva culture medium (UCM) per well.
The UCM consisted of artificial seawater composed of 33 g L−1 of sea salt (Instant Ocean, Aquarium Systems, UK) enriched with 0.1 g L−1 Guillard's F/2 medium (Cell-Hi F2P, Varicon Aqua, UK) and supplemented up to 20 mgNL−1 of NaNO3 (Merk, Germany) to mimic the usual N and P levels in water in a recirculating fish culture system (N = 21.7 ± 2.5 mg L−1 and P = 0.59 ± 0.09 mg L−1, N:P = 80.0 ± 20.8) (Oca et al. 2019). UCM was sterilised using membrane filtration of 0.22 μm (Sterile Cellulose Nitrate, Chmlab Group, Spain).
In the scale-up experiment, sterile (autoclaved) natural seawater from the Vigo estuary, instead of artificial seawater, was used to prepare UCM in the 5-L balloon flasks, while membrane filtered (up to 5 µm) natural seawater was used for UCM in the 40-L tanks.
In the infection assay, 12 discs of a 6-day-old U. ohnoi plate culture not colonised with the probiotic or previously colonised with Phaeobacter sp. (105 CFU cm−2), were placed in the Algae Tank. The Algae Tanks were subjected to aeration and illumination with an intensity of 275 μmol phptons m−2 s−1 (Daylight 6000 K LED panel) and a photoperiod of 12L:12D.
Bacterial inoculum culture
Phaeobacter sp. strain 4UAC3 was isolated from blades of Ulva australis collected in Tragobe (Cambados), Bay of Arousa, Galicia, Spain (42º31′8"N—8º49′7''W), and selected based on antagonism activity against Vibrio anguillarum (unpublished). Phaeobacter sp. 4UAC3 strain was kept stored at -80 °C in Marine Broth (MB, Marine Broth 2216, BD-Difco, Spain) and 15% glycerol (Vorquímica S.L., Spain). The strain was cultured following a protocol described in Prol-García et al. (2009) and the algae cultures were inoculated with Phaeobacter sp. at a final concentration of 107 CFU mL−1. Phaeobacter concentration was estimated by colony forming units (CFU) in Marine Agar (MA, Marine Agar 2216, BD-Difco, Spain) plates. The colonies of Phaeobacter sp. were clearly identifiable due to the formation of brown colonies (Bruhn et al. 2005) on nutrient-rich iron-containing medium as a result of the precipitation of a brown TDA-iron complex (D’Alvise et al. 2016). All medium cultures were previously sterilised in an autoclave at 121 ºC and for 20 min.
Vibrio anguillarum 90–11-287 (Skov et al. 1995) from Instituto de Investigaciones Marinas-Consejo Superior de Investigaciones Cientificas (IIM-CSIC, Spain) collection was used and pre-cultured for 48 h and cultured for 24 h in Marine Broth, as described Prol-García et al. (2009).
Fish-algae cultures
Newly hatched larvae (day 0) of turbot were obtained from Insuíña S.L. (Mougás, Galicia, Spain). After their arrival, they were stored in a tank with 40 L of seawater (filtered and UV sterilised), in the absence of light and acclimatised at ambient temperature.
Three-days old turbot larvae (35 larvae L−1) were placed in the Larvae Tank of the miniaturise IMTA-RAS (Fig. 1) in "green water" with added microalgae (2 × 105 cells of Isochrysis galbana mL−1), moderately aerated (> 90% oxygen saturation) at 20 ºC. The Larvae Tanks were kept under continuous low light (0.8 μmol photons m−2 s−1 intensity on water surface).
Larvae were fed rotifers (Brachionus plicatilis) daily from day 3 to day 10 post-hatch at a final concentration of 5 rotifers per mL, and the seawater was partially renewed (30–40%) every 2 days with the addition of 250 mL of I. galbana culture (2 × 107 cells mL−1) to serve as food for the rotifers as well as to slightly cloud the water to protect the turbot larvae from light. The system was siphoned with sterilised material (by bleach bath) each day to clean the bottom and remove dead larvae for counting.
Infection of larvae with V. anguillarum
To determine the probiotic effect of Ulva with Phaeobacter sp., turbot larvae were challenged with V. anguillarum in an infection model based on bioencapsulation of the pathogen in B. plicatilis, described by a previous protocol (Planas et al. 2005, 2006). The concentration of V. anguillarum in the washed rotifers was 103–104 CFU rotifer−1. The larvae fed with V. anguillarum-loaded rotifers on days 4, 6 and 8 post-hatching.
Sampling of the seaweed-associated bacterial community
Algae discs, previously washed 3 times with 10 mL of SSW, were swabbed on one side with a sterile cotton swab with an applicator stick for 1 min in a laminar flow hood. For the scale-up experiment, the blades taken from the flasks and tank cultures were cut under sterile conditions in 4 cm diameter discs and swabbed as previously described.
The swab was then immersed in a microcentrifuge tube containing 1 mL of SSW, vortexed for 2 min and cotton swab removed. Serial dilutions were made and plated onto MA medium plates. Only plates with 30 and 300 CFU after 3 days of incubation were considered for analysis. Total bacteria were counted including the specific brown colonies representing the inoculated Phaeobacter. The remaining volume of the initial suspension was centrifuged at 13,200 ×g for 15 min and the resulting microbial pellet stored at -20 ºC prior to DNA extraction (see below).
Molecular analysis
DNA extraction
The DNA from bacterial pellets of U. ohnoi was extracted using Instagene Matrix (Bio-Rad Laboratories, USA) according to the manufacturer´s instructions. The DNA was measured in the Nanodrop One (Thermo Scientific, USA) to determine the quantity (ng μL−1) and the quality of DNA, using the absorbance ratio values at A260/A280 and A260/A230. The DNA was stored at -20 °C for the following molecular analysis.
16S rRNA gene amplification
The primers used were two sets of primers targeting the V3-V4 variable region of the 16S rRNA gene. These primers named NOCHL_F (5' TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG 3') and NOCHL_R (5' GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GCM GGG TAT CTA ATC CKG TT 3'). Reactions were performed in a final volume of 25 μL containing 1 μL (10 µM) of each primer, 1.25 μL of Purified BSA (10 mg mL−1, BioLabs, New England Biolabs, UK), 2 μL of DNA template, 7.25 μL Nueclease Free Water (BioLabs, New England Biolabs, UK) and 12.5 μL EconoTaq DNA Polymerase PLUS Green 2X Master Mix (Lucigen Corp, USA). In all reaction, a positive control (2 μL of E. coli DNA) and a negative control (2 μL of nuclease free water) were included. The PCR program consisted of an initial HOT START period at 95 ºC for 2 min. After this, primers were added to each tube and a denaturalization period at 94ºC for 2 min was set, followed by 35 cycles consisting of denaturation at 94 ºC for 30 s, annealing at 50 ºC for 30 s, extension at 72 ºC for 40 s, and a 7 min final extension step at 72 ºC. PCR products were checked by electrophoresis.
16S rRNA gene sequencing and analysis
The PCR products were subjected to another short PCR of 8 cycles using the primer pair 341F/785R (Thijs et al. 2017) containing the Illumina overhang adapter sequences (Kozich et al. 2013). After checking the PCR product by electrophoresis, they were sequenced using the 2 × 300 bp chemistry on Illumina MiSeq sequencer at the Ramaciotti Centre for Genomics (UNSW, Australia).
Sequence processing was performed in RStudio version 4.0.1 (RStudio Team 2020). The raw sequences were processed using the protocol described in (Granzow et al. 2017) and (Mangott et al. 2020). In brief, Trimmomatic (version 0.38) (Bolger et al. 2014) was used to filter the sequencing data according to its quality. This program truncated low quality reads when quality was below 15 in a sliding window of 4 bp, and those shorter than 100 bp. USEARCH (version 11.0.667) was used to process datasets, where paired-end reads were merged (only merging above 80% were considered) and quality filtered. Filtering included the removal of reads shorter than 370 bp or longer than 440 bp, as well as the removal of low-quality reads (expected error > 1) and reads with more than one ambitious base (Wemheuer et al. 2017). Primer sequences were removed and a dereplication process was implemented to obtain unique sequences. Processed sequences of all samples were joined and clustered in Amplicon Sequence Variant (ASVs) using UNOISE algorithm implemented in USEARCH, which de-noise them and generate unique sequences. A de novo chimera removal was added during the clustering step. All ASVs consisting of one single sequence (singletons) were removed and the remaining chimeric sequences were removed using the UCHIME2 algorithm (high confidence mode) (Edgar 2016). To assign taxonomy, ASV sequences were classified using a Bayesian taxonomic classification method BLCA (Gao et al. 2017) (default settings) against the genome taxonomy database (GTDB 207.0) (Parks et al. 2020; Chaumeil et al. 2022).
Data and statistical analysis were conducted in RStudio version 4.0.1. Sampling efficiency and sequencing depth were analysed using rarefaction curves (vegan R package) and the Good's coverage index (QsRutils R package). Count data were normalized using the DESeq2 package to account for uneven sequencing depth between samples (Love et al. 2014). Normalized data were used in measurements of richness and diversity, as well as in distance-based analyses and plots. All other statistical models were carried out using raw, non-transformed data, with the uneven sequencing depth being incorporated in the models.
Previous to performing the statistical analysis, the ASV corresponding to Phaeobacter 4UAC3 was identified and removed from the data set. Firstly, the V3-V4 region was extracted from the complete 16S rRNA sequence of Phaeobacter 4UAC3 previously sequenced (Supplementary Table 1) (unpublished). The V3-V4 region was compared with the ASV.fasta files using the RStudio "blastn" function, indicating that the best performing ASV was ASV6 (100% identity and 100% alignment length).
To analyse the community structure, Bray–Curtis similarity coefficients were calculated using normalized abundances of the ASV sequences (square root transformed data) and the resulting similarity matrix visualized using non-metric multi-dimensional scaling (nMDS). Statistical tests were performed using treatment (Phaeobacter 4UAC3 inoculation and control) and cultivation method (plate, flask and tank) as fixed factors. The effects of each factor on the overall community composition were assessed with permutational multivariate analysis of variance (PERMANOVA) using Bray–Curtis similarity with 999 random permutation (adonis function in the R “vegan” package) (Anderson 2001). Computes Permutational Multivariate Analysis of Dispersion (PERMDISP) was run alongside PERMANOVA to determine if the dispersion of the data has an effect instead of the factors analysed (Warton et al. 2012).
Measures of alpha diversity including unique sequence richness and Inverse Simpson's diversity index were calculated using the vegan R package (Oksanen et al. 2022). Analysis of variance (ANOVA) followed by Tukey's pairwise comparison test was used to assess statistical significance (p < 0.05) between groups adjusted by Benjamini–Hochberg Procedure.
Algae growth parameters
Wet weight of algae was measured by drying the Ulva discs (from plates or flasks) with Whatman paper and weighing each disc on a precision balance (SCALTEC SBA 32, Scaltec, Spain). For the cultures in tanks, Ulva biomass was dried with a salad spinner (2 times 30 s) and then dried with absorbent paper until the paper was dry before weighting. The surface area of the discs was calculated from measurements of the diameters.
The specific growth rate (SGR) was calculated considering the wet weight, with the following formula:
where X1 equates to the wet weight at time one (t1) and X2 represents the wet weight at time two (t2).
Physicochemical analysis and bacterial counts of water
Total bacteria and Phaeobacter monitoring in the water culture of the Ulva was performed using marine agar plates. 1 mL of the culture medium was taken on each sampling day and serial dilutions were performed as described in the previous section "Sampling of the seaweed-associated bacterial community".
The pH of the medium was measured with a pH meter (Sension + pH3, HACH, USA). Salinity was measured with a salinity refractometer (Diesella, UK). Nitrates were measured following the APHA Method 4500-NO3 (Baird et al. 1992)., Briefly, this measurement method is based on the absorbance of the nitrate ion in an aqueous sample, previously filtered through a 0.45 µm filter, at 220 nm and considering an error of the absorbance collected at 275 nm given by the organic matter. The absorbance was measured by using an UV spectrophotometer.
Growth and mortality curves statistics
To check if the presence of Phaeobacter affected the growth of the algae in the scale-up experiment, firstly the two growth curves of the algae (control vs inoculation) were compared by means of the "CGGC permutation test" using the function "compareGrowthCurves" of the R package "statmod" (http://bioinf.wehi.edu.au/software/compareCurves) and adjusting to 1000 permutations. Secondly, the homoscedasticity of the biomass data was checked with a Bartlett's test, and the biomass data at day 21 (the highest growth rate period) were compared using a t-test.
Also, "CGGC permutation test" was implemented between the mortality curves of the 6 experimental cases of the infection assay.
Results
Colonisation and maintenance trial of Phaeobacter sp. on Ulva ohnoi
Algae growth
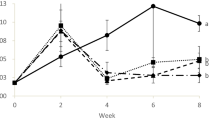
During the first 21 days, the algae experienced exponential growth, reaching a maximum in the SGR between days 14 and 21 of 32.35 ± 3.36% and 36.37 ± 4.26% in UC and UP respectively (Fig. 2). Between days 21 and 28 the growth of the algal thalli in the tanks stagnated, even the SGR decreased. The highest biomass was observed for the UP condition with a maximum of 247 g at 28 days. At that time, part of the biomass was harvested and the tanks restocked at 120 g, which corresponded approximately with the half of the total biomass, to maintain stocking density at similar levels (around 3 kg m−3) and left to grow an additional week. However, the SGR of U. ohnoi between 28 and 35 days was low, similar to the one observed between 21 and 28 days and this fact was not related with depletion of nutrients, as the medium was renewed weekly.
Monitoring of growth and measurement of Ulva biomass in the different experimental conditions in scale-up experiment. a) U. ohnoi without inoculation (UC), b) U. ohnoi with Phaeobacter sp. inoculation (UP). Each point represents an average (n = 2) of the wet weight (g) and Specific Growth Rate (SGR, %) of the sampled algae thalli. Error bars indicate the standard deviation, in those cases were error bars are not seen, it is due to the size of the figure point. The grey dashed lines separate the different culture forms used in the scale-up: 10 mL multi-well plates, 4 L balloon flasks and 40 L tanks. On day 28, the biomass of the culture was reduced (indicated by the black dashed line) to maintain optimal Ulva densities for tank culture
Despite observing a trend which indicates that algae inoculated with Phaeobacter show higher growth, the CGGC permutation test did not find support for significant differences between the algal growth curves of the two treatments (t = -3.200, p = 0.165), and the comparison at day 21 did not indicate significant differences between control and inoculated algae (t = -2.729, df = 2, p = 0.112).
Bacterial counts and bacterial community analysis
Analysis of the culturable bacteria on the algae surface (Fig. 3) showed that although the thallus was treated with sodium hypochlorite to reduce epiphytic bacteria, this treatment did not completely eliminate bacterial growth, with a total bacterial density of 3.85 ± 0.81 Log10 (CFU cm−2) present on the surface at day 0. This density increased slightly at the early stages and in the UC cultures was maintained at values of 3.87 – 6.11 Log10(CFU cm−2). This initial increase in total bacteria was higher in the case of UP cultures, due to the addition of Phaeobacter sp., and remained constant throughout the scaling up process at values between 5.04 – 5.90 Log10(CFU cm−2). We did not observe Phaeobacter in any of the UC cultures.
Total bacteria and Phaeobacter on the surface of U. ohnoi thalli along the scale-up on different treatments in scale-up experiment. a) U. ohnoi without inoculation (UC), b) U. ohnoi with Phaeobacter sp. inoculation (UP). Each point represents an average (n = 2) of the logarithm in base 10 of the concentration of total bacteria and Phaeobacter per surface area of the sampled algae. Error bars indicate the standard deviation. The dashed lines separate the different culture forms used in the scale-up: 10 mL multi-well plates, 4 L balloon flasks and 40 L tanks. Results are based on the CFU counting on Marine Agar plates
In the UP cultures it was observed that, in accordance with the results show in MA, Phaeobacter sp. rapidly colonises the algal surface, reaching 4.90 ± 0.32 Log10(CFU cm−2) after 48 h. Phaeobacter was maintained on the surface of the alga throughout the experiment at a concentration between 4.07 – 5.44 Log10(CFU cm−2). Moreover, during the final phase of the tank experiments (i.e., when the bacteria were no longer re-inoculated), Phaeobacter concentration maintains stable in Ulva until day 21, thereafter the concentration declined from 4.07 ± 1.14 Log10(CFU cm−2) to 1.90 ± 1.25 Log10(CFU cm−2) at 35 days, with a great variability between replicate tanks.
The analysis of the tank water (from day 14 to 35) showed an initial concentration of total bacteria in MA of 5.99 ± 0.28 Log10(CFU cm−2) and 6.29 ± 0.07 Log10(CFU cm−2) in UC and UP respectively. This concentration was reduced at day 28 to 5.29 ± 0.10 Log10(CFU cm−2) and 5.67 ± 0.34 Log10(CFU cm−2), respectively (Fig. 4). The added Phaeobacter reached a concentration of 5.39 ± 1.06 Log10(CFU cm−2) in the water after inoculation, decreasing to 4.12 ± 0.58 Log10(CFU cm−2) at day 21 and was no longer detected by day 28 and 35.
Total bacteria and Phaeobacter in the medium culture of U. ohnoi along the scale-up on different treatments in scale-up experiment. a) U. ohnoi without inoculation (UC), b) U. ohnoi with Phaeobacter sp. inoculation (UP). Each point represents an average (n = 2) of the logarithm in base 10 of the concentration of total bacteria and Phaeobacter per mL of the sampled medium. Error bars indicate the standard deviation. The dashed lines separate the different culture forms used in the scale-up: 10 mL multi-well plates, 4 L balloon flasks and 40 L tanks. Results are based on the CFU counting on Marine Agar plates
Overall, results from the 16S rRNA gene analysis conducted on the bacterial community associated with the surface of U. ohnoi, showed a high taxonomic variability between replicates (Supplementary Fig. 1, 2 and 3), making it difficult to establish a clear grouping of the samples (Supplementary Fig. 1). Statistical analysis was performed with samples from days 7, 14 and 21, corresponding to the end of 7-days periods in each step (plates, flasks and tanks, respectively) in order to study whether the communities would change between cultivation method and Phaeobacter inoculation treatment, using the same time period. PERMANOVA results indicated that only the cultivation method was statistically significant (p-value < 0.05) while treatment did not show to have an effect (p > 0.05) (Supplementary Table 2). PERMDISP also confirmed that the dispersion of the data was not affected by treatment and cultivation method (p > 0.05). Alpha diversity results (Supplementary Fig. 4 and Supplementary Fig. 5) showed that only richness was significantly different between the two treatments and also between the three cultivation methods (p < 0.05), however, a t-test performed between treatment and control groups in each cultivation method, did not show significance (p > 0.05) (Supplementary Table 3).
Phaeobacter inoculum was detected in the 16S rRNA gene sequencing analysis of the Ulva-Phaeobacter condition at days 7, 14 and 21, which corresponds to one week after the inoculation in each step (plate, flask and tank, respectively), with relative abundances of 4.66 ± 6.55%, 1.76 ± 2.50% and 10.35 ± 12.53%, respectively, but showing a high variability between samples (Fig. 5). Phaeobacter was also found in UC case samples with a relative abundance below 0.025 ± 0.022% (Fig. 5).
Box plot of relative abundance in percentage at ASV level of inoculum Phaeobacter from scale-up experiment. Control = Ulva Control case (no inoculation), Inoculation = Ulva Phaeobacter Case (with inoculation), 7 14 21 28 35 = Sampling days, colours = Cultures type (Plate 10 mL, Flask 4 L and Tank 40 L). Results are based on the 16S rRNA gene sequencing
Medium physicochemical analysis
The pH and salinity (Supplementary Fig. 7 and Supplementary Fig. 7, respectively) had similar values in the control (UC) and in the inoculation (UP) treatments, both parameters tended to increase over time due to the photosynthetic activity for pH and evaporation for salinity. The highest pH values were reached in the tanks, while the highest salinity values were reached in the flasks. In the plates the mean pH values were 8.86 ± 0.40 and 8.66 ± 0.33 for UC and UP respectively. In the flasks they were 9.15 ± 0.24 and 9.20 ± 0.21 and in the tanks 9.42 ± 0.37 and 9.40 ± 0.37. Salinity mean values were similar, only slightly higher in the flasks (36.63 ± 2.30 g NaCl L−1 and 37.75 ± 1.06 g NaCl L−1) probably due to a higher evaporation promoted by the aeration system.
Regarding nitrate (Supplementary Fig. 8), it was observed that during algae culture on plates, the concentration of the nutrient decreased drastically in the first two days for both experimental conditions, reaching values of 4.64 ± 1.27 mg N-NO3 L−1 and 4.15 ± 0.04 mg N-NO3 L−1 in the case of UC and UP, respectively. In flasks nitrate concentrations decreased at day 14 from 20.29 ± 0.22 to 5.18 ± 0.59 mg N-NO3 L−1 in the case of UC and from 20.76 ± 0.11 to 8.13 ± 0.16 mg N-NO3 L−1 in the case of UP. In the tanks the nitrate concentrations were kept close to 20.00 mg N-NO3 L−1, and the overall consumption was lower staying above 10.00 mg N-NO3 L−1 for both experimental cases. The consumption of nitrates was related with the density of the culture, being higher in the plates than in the flasks and tanks.
In-vivo antagonism test with turbot larvae (Scophthalmus maximus)
Larvae mortality
Despite a high overall mortality and a large variability, the results obtained showed a positive effect of the presence of U. ohnoi colonised by Phaeobacter sp. (Fig. 6). Indeed, the UP-V condition had at day 4 a lower cumulative mortality (78.9 ± 14%) than the infection control condition (C-V) and the infected Ulva control condition (UC-V) (97.9 ± 1% and 92.9 ± 1% respectively). At the end of the experiment (day 7), the case UP-V showed a mortality of 87.1 ± 14% lower than the other infected conditions. A positive effect on larvae survival was also observed in the situation without Vibrio infection, comparing UP and C. Despite the observed effect, the CGGC permutation test did not find statistical support for significant differences between the six experimental cases, most likely due to the large variability observed between replicates (Supplementary Table 4).
Cumulative mortality of turbot larvae in the different experimental conditions in infection experiment. a) Case control with no infection with Vibrio anguillarum and b) Case infected with Vibrio anguillarum. It its show the average of mortality records (%) (n = 2) in the Larvae Tank of the IMTA-RAS system. The experimental conditions were: Without infection: Control (C), Ulva-Control (UC) and Ulva-Phaeobacter (UP) and with infection with V. anguillarum: Control-Vibrio (C-V), UC-Vibrio (UC-V) and UP-Vibrio (UP-V). Error bars indicate the standard deviation
Bacterial counts analysis
The microbiological analysis of the algae (Supplementary Fig. 9) showed that the initial density of Phaeobacter sp. on Ulva surface was 5.70 ± 0.16 Log10(CFU cm−2). Then, on day 7, Phaeobacter sp. was found with an average density of 4.22 ± 0.66 Log10(CFU cm−2) in UP condition and 4.76 ± 0.43 Log10(CFU cm−2) in UP-V. The initial total bacterial density on the surface of the Ulva discs was 5.90 ± 0.06 Log10(CFU cm−2), increasing it concentration almost 1 Log10(CFU cm−2) at the end of the experiment in all conditions.
At day 6 (Supplementary Fig. 10) the concentration of Phaeobacter in the water was close to 4 Log10(CFU mL−1) in both UP and UP-V cases. Total bacterial counts were close to 5 Log10(CFU mL−1) in the uninfected conditions (being slightly higher in the UP case due to Phaeobacter inoculation), while in the infected cases they were close to 6 due to the addition of V. anguillarum.
Discussion
Colonisation of Phaeobacter sp. 4UAC3 on Ulva ohnoi
The present work investigated the colonisation and maintenance of Phaeobacter sp. 4UAC3 on U. ohnoi with the goal of using Phaeobacter sp. 4UAC3 as a strategy for disease control in fish-algae IMTA-RAS. The aim was to explore the use of Ulva not only as a natural biofilter, which reduces the concentration of nutrients such as nitrates or phosphates in the system water, but also considering it as an holobiont, which includes beneficial microbial communities that could help to maintain the health of the aquaculture system by increasing its resilience to eventual infections of pathogens (e.g. Vibrio spp.).
The scale-up experiment showed the ability of Phaeobacter to colonise and be maintained on the surface of the alga in a large scale for at least two weeks. Maximal growth rate of the alga was observed in the tanks between 14 and 21 days, at which time a harvest could be made. A slight increase in U. ohnoi biomass production was observed when Phaeobacter sp. 4UAC3 is present, but there was no support for statistically significant differences. Positive effects of epiphytic bacteria on the growth and morphology of Ulva species have been reported (Egan et al. 2013). For example, Maribacter spp. induce growth and morphogenesis in axenic Ulva mutabilis (Weiss et al. 2017). On the other hand, it has been reported that co-culture of Phaeobacter spp. (P. gallaeciensis and P. inhibens) with microalgae species can promote the growth of microalgae (Morris et al. 2022; Sauvage et al. 2022). Therefore, the possible positive effect of Phaeobacter sp. 4UAC3 on U. ohnoi should be further investigated and the underlying mechanisms elucidated.
In terms of microbial dynamics Phaeobacter sp. is able to rapidly colonise the surface of the Ulva (within two days) and maintains in constant levels for 21 days. Interestingly, 16S rRNA gene sequencing showed that the relative abundance of Phaeobacter in the inoculated Ulva (Fig. 5) at days 7, 14 and 21, which corresponds to one week after the inoculation in each step (plate, flask and tank, respectively), was higher in tanks, than in flasks and plates, although large variability was observed. This fact could be related to the culture conditions (e.g. light intensity and agitation). Plates are gentle agitated and have the lowest light intensity (115 μmol photons m−2 s−1) and the algal discs received constantly that intensity. In the glass flasks intensity is higher (170 μmol photons m−2 s−1) and due to the shape and volume light attenuation or shading by the Ulva discs can be considered negligible. In contrast, light in the opaque tanks, with illumination on the surface, is influenced by depth and the shading resulting from algae density (Oca et al. 2019) and by chlorophyll content (Masaló et al. unpublished). Recent results in our laboratory have shown that light intensity affects the maintenance of Phaeobacter biofilms on Ulva surfaces (unpublished) a phenomenon warranting future investigation.
In our experimental conditions Phaeobacter did not out-compete the natural microbiota present in U. ohnoi surface. Competitive exclusion by Phaeobacter gallaeciensis has been demonstrated by Rao et al. (2010) in U. australis biofilms in axenic algae discs previously colonised by other bacteria (e.g. Pseudoalteromonas tunicata). However, in non-axenic Ulva, the preestablished multi-species biofilms would be more resilient and therefore resist displacement or antagonism by other bacteria. In the present work Phaeobacter was showed to colonise the U. ohnoi surface without significantly modifying the bacterial communities, as observed in the 16S rRNA gene diversity analysis (Supplementary Fig. 2 and 3). Rao et al. (2006) suggest that Phaeobacter may take advantage of its ability to use a wide range of carbon sources (citing Ruiz-Ponte et al. (1998)), which can enable this bacterium the access to nutrients and to colonize preestablished biofilms.
Moreover, Phaeobacter spp. were not predominant in the Ulva species sampled in Galicia (U. rigida and U. australis) from which Phaeobacter 4UAC3 was isolated (Roth-Schulze et al. 2018); NCBI http://www.ncbi.nlm.nih.gov/Traces/ SRA Accession ID SRP087427). This would suggest that although Phaeobacter spp. are generally present in Ulva species, they are not abundant and their abundance is enhanced only under certain circumstances. This hypothesis is aligned with the idea that marine macroalgae favour the maintenance of low abundant marine microbial diversity, i.e. “sustaining the rare” (Troussellier et al. 2017), that become abundant only in the appropriate environmental conditions. Ulva in IMTA-RAS aquaculture systems with a high nutrient load may provide favourable conditions to Phaeobacter and its predominance when inoculated.
When re-inoculated at the beginning of each step of the scale-up Phaeobacter can be maintain on the algae in the tanks until harvesting. Prol-García and Pintado (2013) demonstrated the ability of P. gallaeciensis 27–4 to colonize and maintain on biofilter inert supports (plastic, glass or ceramic). In the present work the maintenance is not only due to the ability of the bacteria to adhere to the laminar surface of the alga, but also to the possible contribution of nutrients as the presence of carbon-rich compounds produced by algae and found on their surface (e.g. agar, carrageenan, alginate, fucans, ulvans, laminarin, cellulose or pectin) is an important factor for bacterial colonisation (Egan et al. 2013).
It should be noted that after the last re-inoculation (day 14), Phaeobacter numbers remain at a constant level for two weeks (until day 28) before decreasing sharply and becoming almost undetectable at day 35. The bacterial diversity analysis from 16S rRNA sequencing indicated that Phaeobacter sp. did not alter the microbial communities on the Ulva surface, as no significant differences were found between treatments and no clear clustering was observed in the nMDS plot. However, a study with more replicates will be needed to confirm those results as Ulva species show a high taxonomic variability in bacterial communities, even among individuals from the same populations (Tujula et al. 2009) keeping in common a core of functional genes (Roth-Schulze et al. 2018).
It was observed that in the inoculation treatment, when Phaeobacter decreases to very low levels (day 35), the microbial communities of Ulva seem to regain their initial conditions in terms of population, and tend to cluster near the initial samples (day 7 and 14) (Supplementary Fig. 1). This fact could be due to some external factor that promotes the disappearance of Phaeobacter sp., as changes in the composition of the medium, or physico-chemical factors on the Ulva surface. It has been documented that Phaeobacter does not proliferate well in alkaline media at pHs above 9.5, being the optimum growth between 7.0–8.0 (Breider et al. 2017; Martens et al. 2006). Increasing the light in successive steps of the scaling (115 μmol photons m−2 s−1, 170 μmol photons m−2 s−1 and 275 μmol photons m−2 s−1) promotes an increase in photosynthetic processes leading to increased growth of the algae and alkalinisation of the medium. In fact, the initial pH in the multi-well plates was ~ 8.5, while in the tanks there were times when the pH value exceeded 9.5. Thus, it would be interesting to investigate the effect of these parameters (light intensity, algal growth, and pH) on the maintenance of Phaeobacter on the algae.
During culture in the tanks, Phaeobacter sp. is released from the surface of U. ohnoi and passes into the water, reaching levels of 104–105 CFU mL−1 at 14 and 21 days, respectively. These levels in water are somewhat higher than those obtained by Prol-García et al. (2014) with inert biofilters (about 103 CFU· mL−1), which proved to be effective, inhibiting and inactivating V. anguillarum and V. splendidus. This is interesting because for an application in IMTA-RASs it would make it possible for the probiotic to reach the fish tank and exert its antagonistic activity, a fact that has been verified in the larval infection trials. Nevertheless, Phaeobacter sp. disappears from the water after 21 days, as it also diminishes on the surface of the alga. It is assumed that from this moment onwards, its antagonistic effect on the water of the systems will decrease.
Thus, in an IMTA-RAS, a harvest can be envisaged every 21 days. In order to sustain the culture of the algae and the probiotic effects on the fish larvae, the new culture could be started with algae previously colonised by Phaeobacter in the tanks from which the biomass was taken. However, a protocol to maintain a constant colonization of the system, by adjusting the variables that may affect the permanence of Phaeobacter on Ulva, should be further explored.
In-vivo antagonistic effect in larval fish culture
This study demonstrated the feasibility of co-culturing U. ohnoi with Phaeobacter bacteria and also indicates the possibility of using this approach as a strategy to control vibriosis in fish-algae IMTA-RAS. In a challenge model with turbot larvae in a miniaturised IMTA-RAS, U. ohnoi colonised with Phaeobacter sp. a reduction of mortality in larvae experimentally infected with V. anguillarum was observed, although this trend was not statistically significant.
In the experimental challenge model used with fish larvae, V. anguillarum was introduced by prey (rotifers), simulating the usual route of entry into a turbot larvae culture. Moreover, from this host, V. anguillarum can transfer to the water (Prol-García et al. 2009). Planas et al. (2006) showed that V. anguillarum did not colonise the intestines of turbot larvae but infected the epidermis of the larvae, and it was suggested that the presence of Phaeobacter in tank water would antagonise its detrimental effect on the larvae. In our study, the presence of Phaeobacter sp. 4UAC3 on the surface of the algae improved larval survival by almost 20% (at day 4) during V. anguillarum infections in the small-scale IMTA-RAS. This increase in survival, although not significantly supported by statistical tests, is in accordance with that obtained by Prol-García and Pintado (2013) in a similar in vivo test but in a closed tank, without recirculation. However, the in-vivo experiment in the miniaturised IMTA-RASs showed a general higher larvae mortality than the ones in closed tanks (Prol-García and Pintado 2013), and due to a high variability statistical analysis did not support for significant differences between treatments. Therefore, it will be necessary to optimise a test in IMTA-RASs to achieve lower mortality and better measure the probiotic effect. The use of juveniles, instead of larvae, or sole, instead of turbot, should be considered.
Phaeobacter was found in Ulva in the IMTA-RAS at a density similar to the one observed in the scale-up experiment, with an average density at day 7 close to 104 CFU cm−2 among 105 CFU cm−2 total bacteria. This indicated that the probiotic is maintained in high abundance in the IMTA-RASs, in which the nutrient supply conditions were different from those in the algal scale-up experiment. In the water, the concentration of Phaeobacter was close to 103 CFU mL−1 after about 6 days in both experiments. This showed that the colonised Ulva provides a constant supply of the probiotic to the recirculating water in the IMTA-RASs, and that the concentration in water was enough to obtain an effect on Vibrio antagonism.
We can conclude that Phaeobacter sp. 4UAC3 is able to colonise U. ohnoi and maintain itself in a re-inoculation scale-up process from multi-well plates (10 mL) to flasks (4 L) and finally to tanks (40 L). Phaeobacter-colonized U. ohnoi discs could have a positive effect by reducing larvae mortality between 10 and 20%, according to the trends observed in the infection trials on turbot larvae cultured in small-scale IMTA-RASs (Ulva ohnoi—Scophthalmus maximus). However further experiments and test are required to define the exact impact and benefit that Phaeobacter-colonized U. ohnoi has on IMTA-RASs. These results indicated that Ulva species colonised with Phaeobacter sp. 4UAC3 could be used in IMTA-RASs as an alternative to traditional disease control methods. However, a reduction of Phaeobacter sp. 4UAC3 was observed over time and further studies are being conducted to investigate Ulva-Phaeobacter biochemical interactions and the factors that would determine those interactions, with the objective of establishing the conditions and design strategies to increase the maintenance the probiotic on the surface of the algae, and implement this approach to a large scale IMTA-RAS.
Data availability
All raw sequences used in this study have been submitted to the NCBI BioProject database ID: PRJNA921851 under Submission ID SUB12518914 and will be publicly available at http://www.ncbi.nlm.nih.gov/bioproject/921851
Code availability
The code used in this study can be shared upon request.
References
Anderson MJ (2001) A new method for non-parametric multivariate analysis of variance. Austral Ecol 26:32–46
Baird RB, Eaton AD, Clesceri LS (1992) APHA Method 4500-NO3. In: Rice BEW (ed) Standard methods for the examination of water and wastewater, 18th edn. American Public Health Association, pp 87–94
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Bolton JJ, Cyrus MD, Brand MJ, Joubert M, Macey BM (2016) Why grow Ulva? Its potential role in the future of aquaculture. Perspectives in Phycology 3:113–120
Breider S, Freese HM, Spröer C, Simon M, Overmann J, Brinkhoff T (2017) Phaeobacter porticola sp. nov., an antibiotic-producing bacterium isolated from a sea harbour. Int J Syst Evol Microbiol 67(7):2153–2159
Brinkhoff T, Bach G, Heidorn T, Liang L, Schlingloff A, Simon M (2004) Antibiotic production by a Roseobacter clade-affiliated species from the German Wadden Sea and its antagonistic effects on indigenous isolates. Appl Environ Microbiol 70:2560–2565
Bruhn JB, Nielsen KF, Hjelm M, Hansen M, Bresciani J, Schulz S, Gram L (2005) Ecology, inhibitory activity, and morphogenesis of a marine antagonistic bacterium belonging to the Roseobacter clade. Appl Environ Microbiol 71:7263–7270
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH (2022) GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38:5315–5316
Cremades J, Casais E, Oca J, Masaló I, Jimenez P, Pintado J (2017) Ulva ohnoi (Ulvales, Chlorophyta) as a biofilter in IMTA-RAS systems: influence of nitrogen and phosphorous levels in its development. Phycologia 56:36
D’Alvise PW, Lillebø S, Wergeland HI, Gram L, Bergh Ø (2013) Protection of cod larvae from vibriosis by Phaeobacter spp.: A comparison of strains and introduction times. Aquaculture 384–387:82–86
D’Alvise PW, Phippen CBW, Nielsen KF, Gram L (2016) Influence of iron on production of the antibacterial compound tropodithietic acid and its noninhibitory analog in Phaeobacter inhibens. Appl Environ Microbiol 82:502
de Schryver P, Vadstein O (2014) Ecological theory as a foundation to control pathogenic invasion in aquaculture. ISME J 8:2360–2368
Edgar RC (2016) UCHIME2: improved chimera prediction for amplicon sequencing. BioRxiv 074252
Egan S, Gardiner M (2016) Microbial dysbiosis: Rethinking disease in marine ecosystems. Front Microbiol 7:991
Egan S, Harder T, Burke C, Steinberg P, Kjelleberg S, Thomas T (2013) The seaweed holobiont: Understanding seaweed-bacteria interactions. FEMS Microbiol Rev 37:462–476
Gao X, Lin H, Revanna K, Dong Q (2017) A Bayesian taxonomic classification method for 16S rRNA gene sequences with improved species-level accuracy. BMC Bioinformatics 18:247
Granzow S, Kaiser K, Wemheuer B, Pfeiffer B, Daniel R, Vidal S, Wemheuer F (2017) The effects of cropping regimes on fungal and bacterial communities of wheat and Faba bean in a greenhouse pot experiment differ between plant species and compartment. Front Microbiol 8:902
Hayden HS, Blomster J, Maggs CA, Silva PC, Stanhope MJ, Waaland JR (2003) Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. Eur J Phycol 38:277–294
Hollants J, Leliaert F, de Clerck O, Willems A (2013) What we can learn from sushi: A review on seaweed-bacterial associations. FEMS Microbiol Ecol 83:1–16
Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD (2013) Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79:5112–5120
Lawton RJ, Mata L, de Nys R, Paul NA (2013) Algal bioremediation of waste waters from land-based aquaculture using Ulva: selecting target species and strains. PLoS ONE 8(10):e77344
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Mangott A, Nappi J, Delli Paoli Carini A, Goncalves P, Hua K, Domingos JA, de Nys R, Thomas T (2020) Ulva lactuca as a functional ingredient and water bioremediator positively influences the hepatopancreas and water microbiota in the rearing of Litopenaeus vannamei. Algal Res 51:102040
Manhart JR (1994) Phylogenetic analysis of green plant rbcL sequences. Mol Phylogenet Evol 3:114–127
Mantri VA, Kazi MA, Balar NB, Gupta V, Gajaria T (2020) Concise review of green algal genus Ulva Linnaeus. J Appl Phycol 32:2725–2741
Martens T, Heidorn T, Pukal R, Simon M, Tindall BJ, Brinkhoff T (2006) Reclassification of Roseobacter gallaeciensis Ruiz-Ponte et al. 1998 as Phaeobacter gallaeciensis gen. nov., comb. nov., description of Phaeobacter inhibens sp. nov., reclassification of Ruegeria algicola (Lafay et al. 1995) Uchino et al. 1999 as Marinovum algicola gen. nov., comb. nov., and emended descriptions of the genera Roseobacter, Ruegeria and Leisingera. Int J Syst Evol Microbiol 56:1293–1304
Morris MM, Kimbrel JA, Geng H, Tran-Gyamfi MB, Yu ET, Sale KL, Lane TW, Mayali X (2022) Bacterial community assembly, succession, and metabolic function during outdoor cultivation of Microchloropsis salina. MSphere 7(4):e0023122
Oca J, Cremades J, Jiménez P, Pintado J, Masaló I (2019) Culture of the seaweed Ulva ohnoi integrated in a Solea senegalensis recirculating system: influence of light and biomass stocking density on macroalgae productivity. J Appl Phycol 31:2461–2467
Oksanen J, Gavin L, Simpson F, Blanchet G, Kindt R, Legendre P et al. (2022) Package ‘Vegan’ Community Ecology Package. https://CRAN.R-project.org/package=vegan
Parks DH, Chuvochina M, Chaumeil PA, Rinke C, Mussig AJ, Hugenholtz P (2020) A complete domain-to-species taxonomy for Bacteria and Archaea. Nature Biotech 38:1079–1086
Planas M, Pérez-Lorenzo M, Hjelm M, Gram L, Uglenes Fiksdal I, Bergh Ø, Pintado J (2006) Probiotic effect in vivo of Roseobacter strain 27–4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 255:323–333
Planas M, Pérez-Lorenzo M, Vázquez JA, Pintado J (2005) A model for experimental infections with Vibrio (Listonella) anguillarum in first feeding turbot (Scophthalmus maximus L.) larvae under hatchery conditions. Aquaculture 250:232–243
Prol-García MJ, Bruhn JB, Pintado J, Gram L (2009) Real-time PCR detection and quantification of fish probiotic Phaeobacter strain and fish pathogenic Vibrio in microalgae, rotifer, Artemia and first feeding turbot (Psetta maxima) larvae. J Appl Microbiol 106:1292–1303
Prol-García MJ, Gómez M, Sánchez L, Pintado J (2014) Phaeobacter grown in biofilters: a new strategy for the control of Vibrionaceae in aquaculture. Aquacult Res 45:1012–1025
Prol-García MJ, Pintado J (2013) Effectiveness of probiotic Phaeobacter bacteria grown in biofilters against Vibrio anguillarum infections in the rearing of turbot (Psetta maxima) larvae. Mar Biotech 15:726–738
Rao D, Skovhus T, Tujula N, Holmström C, Dahllöf I, Webb JS, Kjelleberg S (2010) Ability of Pseudoalteromonas tunicata to colonize natural biofilms and its effect on microbial community structure. FEMS Microbiol Ecol 73:450–457
Rao D, Webb JS, Kjelleberg S (2006) Microbial colonization and competition on the marine alga Ulva australis. Appl Environ Microbiol 72:5547–5555
Roth-Schulze AJ, Pintado J, Zozaya-Valdés E, Cremades J, Ruiz P, Kjelleberg S, Thomas T (2018) Functional biogeography and host specificity of bacterial communities associated with the marine green alga Ulva spp. Mol Ecol 27:1952–1965
RStudio Team (2020) RStudio: Integrated Development for R. RStudio. PBC, Boston, MA. http://www.rstudio.com/
Ruiz-Ponte C, Cilia V, Lambert C, Nicolas JL (1998) Roseobacter gallaeciensis sp. nov., a new marine bacterium isolated from rearings and collectors of the scallop Pecten maximus. Int J Syst Bacteriol 48:537–542
Sauvage J, Wikfors GH, Dixon MS, Kapareiko D, Sabbe K, Li X, Joyce A (2022) Bacterial exudates as growth-promoting agents for the cultivation of commercially relevant marine microalgal strains. J World Aquacult Soc 53:1101–1119
Skov MN, Pedersen K, Larsen JL (1995) Comparison of pulsed-field gel electrophoresis, ribotyping, and plasmid profiling for typing of Vibrio anguillarum serovar O1. Appl Environ Microbiol 61:1540–1545
Tesdorpf JE, Geers AU, Strube ML, Gram L, Bentzon-Tilia M (2022) Roseobacter group probiotics exhibit differential killing of fish pathogenic Tenacibaculum species. Appl Environ Microbiol 88(6):e0241821
Thijs S, Op De Beeck M, Beckers B, Truyens S, Stevens V, Van Hamme JD, Weyens N, Vangronsveld J (2017) Comparative evaluation of four bacteria-specific primer pairs for 16S rRNA gene surveys. Front Microbiol 8:494
Troussellier M, Escalas A, Bouvier T, Mouillot D (2017) Sustaining rare marine microorganisms: Macroorganisms as repositories and dispersal agents of microbial diversity. Front Microbiol 8:00947
Tujula NA, Crocetti GR, Burke C, Thomas T, Holmström C, Kjelleberg S (2009) Variability and abundance of the epiphytic bacterial community associated with a green marine Ulvacean alga. ISME J 4:301–311
Warton DI, Wright ST, Wang Y (2012) Distance-based multivariate analyses confound location and dispersion effects. Meth Ecol Evol 3:89–101
Weiss A, Costa R, Wichard T (2017) Morphogenesis of Ulva mutabilis (Chlorophyta) induced by Maribacter species (Bacteroidetes, Flavobacteriaceae). Bot Mar 60:197–206
Wemheuer B, Wemheuer F, Meier D, Billerbeck S, Giebel HA, Simon M, Scherber C, Daniel R (2017) Linking compositional and functional predictions to decipher the biogeochemical significance in DFAA turnover of abundant bacterioplankton lineages in the North Sea. Microorganisms 5:68
Wichard T, Charrier B, Mineur F, Bothwell JH, de Clerck O, Coates JC (2015) The green seaweed Ulva: a model system to study morphogenesis. Front Plant Sci 6:72
Acknowledgements
The authors thank, Alexandro Chamorro, technician of aquaria at IIM-CSIC, for the assistance in setting the aquaria systems. We also thank Sara Barwani and Sarah Langhorne, BSc students from the University of Portsmouth who did a Traineeship at IIM-CSIC funded by Eramus+ program that was framed in the development of this work. We thank Mariana Pereira at IIM-CSIC for assistance with the statistical analysis.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. The IntegRas project (AGL2013-41868-R) and the UlvaQuo Project (RTI2018-095062-B-C21) were funded by the Spanish Agencia Estatal de Investigación. Gonzalo Del Olmo was beneficiary of a grant from the Galician Axencia Galega de Innovación (IN606A-2021/038).
Author information
Authors and Affiliations
Contributions
JP, JC and IM obtained funding for the study. JP, JC and PR designed the study. TG, JP and PR performed the experiments. JP, JC, TG, PR and IM participated in the analysis of data. JN, TT, SE and GDO participated in the preparation of samples for sequencing and bioinformatics analysis. JP and GDO wrote the first draft of the manuscript. All authors contributed to the writing of the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no competing interests to declare that are relevant to the content of this article.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pintado, J., Del Olmo, G., Guinebert, T. et al. Manipulating the Ulva holobiont: Co-culturing Ulva ohnoi with Phaeobacter bacteria as a strategy for disease control in fish-macroalgae IMTA-RAS aquaculture. J Appl Phycol 35, 2017–2029 (2023). https://doi.org/10.1007/s10811-023-02986-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10811-023-02986-1