Abstract
Purpose
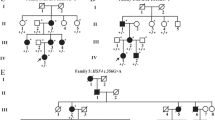
To investigate the potential genetic defects in a five-generation Chinese family with autosomal dominant congenital cataract (ADCC).
Methods
Whole exome sequencing was performed to search the variants in the candidate genes associated with congenital cataract. Sanger sequencing was used to validate the variants and examine their co-segregation in the patients and their relatives. The potential effect of the variants was analyzed using several bioinformatic methods and further examined through Western blotting and co-immunoprecipitation.
Results
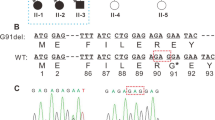
A missense variant c. 71 G > T (p. Gly24Val) in the CRYBA4 gene, a known ADCC candidate gene, was identified to be heterozygously present in the patients and co-segregate with cataract in the family. The mutation was absent in all of the searched databases, including our in-house exome sequences of 10,000 Chinese. The alignments of the amino acid sequences of CRYBA4 in a variety of species revealed that the amino acid residue Gly24 was evolutionarily highly conserved, and the in silico analysis predicted that the missense mutation of Gly24Val was damaging for the protein structure and function of CRYBA4. Then, the in vitro expression analysis further revealed that the Gly24Val mutation in CRYBA4 inhibited its binding with CRYBB1. The impaired interaction of β-crystallin proteins may affect their water-solubility and contribute to the formation of precipitates in lens fiber cells.
Conclusion
We identified a novel missense variant in the CRYBA4 gene as a pathogenic mutation of ADCC in a Chinese family. Our finding expanded the CRYBA4 variation spectrum associated with congenital cataracts.



Similar content being viewed by others
Data availability
The data used to support this study are available from the corresponding author upon request.
References
Robinson GC, Jan JE, Kinnis C (1987) Congenital ocular blindness in children, 1945 to 1984. Am J Dis Child 141(12):1321–1324. https://doi.org/10.1001/archpedi.1987.04460120087041
Apple DJ, Ram J, Foster A, Peng Q (2000) Elimination of cataract blindness: a global perspective entering the new millenium. Surv Ophthalmol 45(Suppl 1):S1–196
Gilbert C, Foster A (2001) Childhood blindness in the context of VISION 2020–the right to sight. Bull World Health Organ 79(3):227–232
Holmes JM, Leske DA, Burke JP, Hodge DO (2003) Birth prevalence of visually significant infantile cataract in a defined U.S. population. Ophthalmic Epidemiol 10(2):67–74. https://doi.org/10.1076/opep.10.2.67.13894
Sheeladevi S, Lawrenson JG, Fielder AR, Suttle CM (2016) Global prevalence of childhood cataract: a systematic review. Eye 30(9):1160–1169. https://doi.org/10.1038/eye.2016.156
Francois J (1982) Genetics of cataract. Ophthalmologica 184(2):61–71. https://doi.org/10.1159/000309186
Haargaard B, Wohlfahrt J, Fledelius HC, Rosenberg T, Melbye M (2004) A nationwide Danish study of 1027 cases of congenital/infantile cataracts: etiological and clinical classifications. Ophthalmology 111(12):2292–2298. https://doi.org/10.1016/j.ophtha.2004.06.024
Hejtmancik JF (2008) Congenital cataracts and their molecular genetics. Semin Cell Dev Biol 19(2):134–149. https://doi.org/10.1016/j.semcdb.2007.10.003
Wirth MG, Russell-Eggitt IM, Craig JE, Elder JE, Mackey DA (2002) Aetiology of congenital and paediatric cataract in an Australian population. Brit J Ophthalmol 86(7):782–786. https://doi.org/10.1136/bjo.86.7.782
Wistow GJ, Piatigorsky J (1988) Lens crystallins - the evolution and expression of proteins for a highly specialized tissue. Annu Rev Biochem 57:479–504. https://doi.org/10.1146/annurev.bi.57.070188.002403
Wistow G (2012) The human crystallin gene families. Hum Genomics 6:26. https://doi.org/10.1186/1479-7364-6-26
Billingsley G, Santhiya ST, Paterson AD, Ogata K, Wodak S, Hosseini SM, Manisastry SM, Vijayalakshmi P, Gopinath PM, Graw GJ, Heon E (2006) CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet 79(4):702–709. https://doi.org/10.1086/507712
Xu J, Zhao WJ, Chen XJ, Yao K, Yan YB (2018) Introduction of an extra tryptophan fluorophore by cataract-associating mutations destabilizes betaB2-crystallin and promotes aggregation. Biochem Biophys Res Commun 504(4):851–856. https://doi.org/10.1016/j.bbrc.2018.09.028
Xi YB, Zhang K, Dai AB, Ji SR, Yao K, Yan YB (2014) Cataract-linked mutation RI 88H promotes beta B2-crystallin aggregation and fibrillization during acid denaturation. Biochem Biophys Res Commun 447(2):244–249. https://doi.org/10.1016/j.bbrc.2014.03.119
Zhu S, Xi XB, Duan TL, Zhai Y, Li J, Yan YB, Yao K (2018) The cataract-causing mutation G75V promotes gammaS-crystallin aggregation by modifying and destabilizing the native structure. Int J Biol Macromol 117:807–814. https://doi.org/10.1016/j.ijbiomac.2018.05.220
Shiels A, Bennett TM, Hejtmancik JF (2010) Cat-Map: putting cataract on the map. Mol Vis 16(215–19):2007–2015
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. https://doi.org/10.1038/msb.2011.75
Ashkenazy H, Abadi S, Martz E, Chay O, Mayrose I, Pupko T, Ben-Tal N (2016) ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res 44(W1):W344–W350. https://doi.org/10.1093/nar/gkw408
Geourjon C, Deleage G (1995) SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput Appl Biosci 11(6):681–684. https://doi.org/10.1093/bioinformatics/11.6.681
Kaplan W, Littlejohn TG (2001) Swiss-PDB Viewer (Deep View). Brief Bioinform 2(2):195–197. https://doi.org/10.1093/bib/2.2.195
Capriotti E, Fariselli P, Casadio R (2005) I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res 33:306–310. https://doi.org/10.1093/nar/gki375
Cheng J, Randall A, Baldi P (2006) Prediction of protein stability changes for single-site mutations using support vector machines. Proteins 62(4):1125–1132. https://doi.org/10.1002/prot.20810
Savojardo C, Fariselli P, Martelli PL, Casadio R (2016) INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics 32(16):2542–2544. https://doi.org/10.1093/bioinformatics/btw192
Fraczkiewicz RW (1998) Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J Comp Chem 19:319–333
Walker JM (2005) The proteomics protocols handbook. Humana Press, Totowa
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7(4):248–249. https://doi.org/10.1038/nmeth0410-248
Choi Y, Chan AP (2015) PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31(16):2745–2747. https://doi.org/10.1093/bioinformatics/btv195
Liu YQ, Lu YJ, Liu SS, Liao SY (2017) Novel compound heterozygous mutations of ALDH1A3 contribute to anophthalmia in a non-consanguineous Chinese family. Genet Mol Biol 40(2):430–435. https://doi.org/10.1590/1678-4685-Gmb-2016-0120
Yi J, Yun J, Li ZK, Xu CT, Pan BR (2011) Epidemiology and molecular genetics of congenital cataracts. Int J Ophthalmol 4(4):422–432. https://doi.org/10.3980/j.issn.2222-3959.2011.04.20
Li FF, Zhu SQ, Wang SZ, Gao C, Huang SZ, Zhang M, Ma X (2008) Nonsense mutation in the CRYBB2 gene causing autosomal dominant progressive polymorphic congenital coronary cataracts. Mol Vis 14:750–755. http://47mv-v14-750-dimer.pdf
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, Committee ALQA (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17(5):405–424. https://doi.org/10.1038/gim.2015.30
Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, Beroud C (2009) Human splicing finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37(9):e67. https://doi.org/10.1093/nar/gkp215
Slingsby C, Clout NJ (1999) Structure of the crystallins. Eye 13:395–402. https://doi.org/10.1038/eye.1999.113
Bateman OA, Sarra R, van Genesen ST, Kappe G, Lubsen NH, Slingsby C (2003) The stability of human acidic beta-crystallin oligomers and hetero-oligomers. Exp Eye Res 77(4):409–422. https://doi.org/10.1016/S0014-4835(03)00173-8
Liu BF, Liang JJ (2007) Protein-protein interactions among human lens acidic and basic beta-crystallins. FEBS Lett 581(21):3936–3942. https://doi.org/10.1016/j.febslet.2007.07.022
Marin Vinader L, Onnekink C, Van Genesen ST, Slingsby C, Lubsen NH (2006) In vivo heteromer formation Expression of soluble betaA4-crystallin requires coexpression of a heteromeric partner. FEBS J 273(14):3172. https://doi.org/10.1111/j.1742-4658.2006.05326.x
Bateman JB, Von-Bischhoffshaunsen FR, Richter L, Flodman P, Burch D, Spence MA (2007) Gene conversion mutation in crystallin, beta-B2 (CRYBB2) in a Chilean family with autosomal dominant cataract. Ophthalmology 114(3):425–432. https://doi.org/10.1016/j.ophtha.2006.09.013
Lubsen NH, Aarts HJ, Schoenmakers JG (1988) The evolution of lenticular proteins: the beta- and gamma-crystallin super gene family. Prog Biophys Mol Biol 51(1):47–76. https://doi.org/10.1016/0079-6107(88)90010-7
Rose GD, Fleming PJ, Banavar JR, Maritan A (2006) A backbone-based theory of protein folding. Proc Natl Acad Sci U S A 103(45):16623–16633. https://doi.org/10.1073/pnas.0606843103
Li W, Ji Q, Wei Z, Chen YL, Zhang Z, Yin X, Aghmiuni SK, Liu M, Chen W, Shi L, Chen Q, Du X, Yu L, Cao MJ, Wang Z, Huang S, Jin T, Wang Q (2019) Biochemical characterization of G64W mutant of acidic beta-crystallin 4. Exp Eye Res 186:107712. https://doi.org/10.1016/j.exer.2019.107712
Graw J (2009) Genetics of crystallins: cataract and beyond. Exp Eye Res 88(2):173–189. https://doi.org/10.1016/j.exer.2008.10.011
Marin-Vinader L, Onnekink C, van Genesen ST, Slingsby C, Lubsen NH (2006) In vivo heteromer formation. Expression of soluble beta A4-crystallin requires coexpression of a heteromeric partner. FEBS J 273(14):3172–3182. https://doi.org/10.1111/j.1742-4658.2006.05326.x
Ecroyd H, Carver JA (2009) Crystallin proteins and amyloid fibrils. Cell Mol Life Sci 66(1):62–81. https://doi.org/10.1007/s00018-008-8327-4
Yu Y, Qiao Y, Ye Y, Li J, Yao K (2021) Identification and characterization of six beta-crystallin gene mutations associated with congenital cataract in Chinese families. Mol Genet Genomic Med 9(3):e1617. https://doi.org/10.1002/mgg3.1617
Acknowledgements
This work was supported by Grants from the National Natural Science Foundation of China (No.81773159).
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number: No.81773159).
Author information
Authors and Affiliations
Contributions
XZ: participated in the whole study, drafting of the manuscript, and data analysis; XZ, CL, ML, XL, ZW, SX and XT: carried out the experiments; YY: contributed to data curation; YL: contributed to writing—review, editing, correspondence, and proofs of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare no conflict of interest. The authors have no relevant financial or non-financial interests to disclose.
Consent to publish
The authors affirm that human research participants provided informed consent for publication of the images in Figure 1b.
Ethical approval
The current study was reviewed and approved by the Research Ethics Committee of the West China Hospital, West China Medical School, Sichuan University (2019–772).
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, X., Liang, C., Liu, M. et al. A novel missense variant c.71G > T (p.Gly24Val) of the CRYBA4 gene contributes to autosomal-dominant congenital cataract in a Chinese family. Int Ophthalmol 43, 43–50 (2023). https://doi.org/10.1007/s10792-022-02386-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10792-022-02386-3